9AUL
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUJ
 
 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
5U6C
 
 | | Crystal structure of the Mer kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase Mer | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
5UWD
 
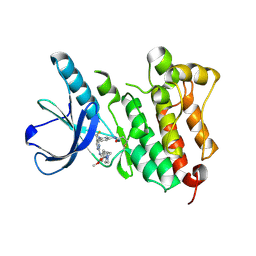 | |
3I6C
 
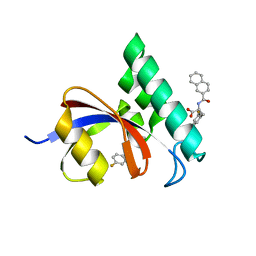 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | 3-fluoro-N-(naphthalen-2-ylcarbonyl)-D-phenylalanine, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3IK8
 
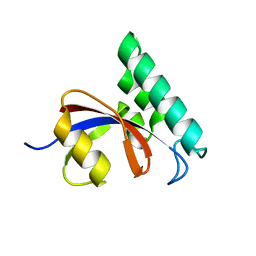 | |
3IKG
 
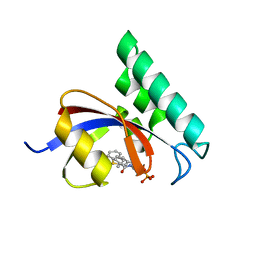 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3RWQ
 
 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Kazmirski, S, Kohls, D. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
4HZS
 
 | |
4HZR
 
 | | Crystal structure of Ack1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, Activated CDC42 kinase 1, CHLORIDE ION, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Ack1: activation and regulation by allostery.
Plos One, 8, 2013
|
|
5U6B
 
 | | Structure of the Axl kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase receptor UFO | | Authors: | Gajiwala, K.S, Grodsky, N. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
5U8L
 
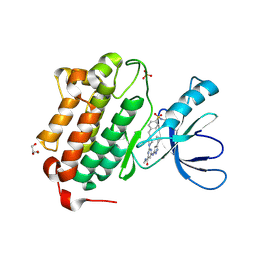 | | Crystal structure of EGFR kinase domain in complex with a sulfonyl fluoride probe XO44 | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
8PK2
 
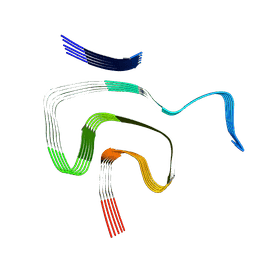 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PIX
 
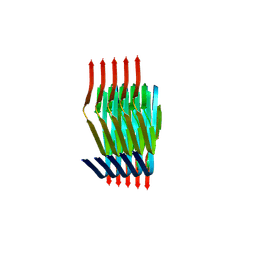 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PJO
 
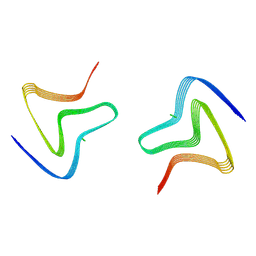 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
9FYP
 
 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
7N8Y
 
 | |
7KCN
 
 | | Reconstructed ancestor of HIUases and Transthyretins | | Descriptor: | 1,2-ETHANEDIOL, HIUase-TTR ancestor, PHOSPHATE ION | | Authors: | Nagem, R.A.P, Bleicher, L, Costa, M.A.F. | | Deposit date: | 2020-10-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Reenacting the Birth of a Function: Functional Divergence of HIUases and Transthyretins as Inferred by Evolutionary and Biophysical Studies.
J.Mol.Evol., 89, 2021
|
|
7KJJ
 
 | | Reconstructed ancestor of HIUases and Transthyretins | | Descriptor: | 1,2-ETHANEDIOL, 3,5,3',5'-TETRAIODO-L-THYRONINE, TTR ancestor | | Authors: | Nagem, R.A.P, Bleicher, L, Costa, M.A.F. | | Deposit date: | 2020-10-26 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reenacting the Birth of a Function: Functional Divergence of HIUases and Transthyretins as Inferred by Evolutionary and Biophysical Studies.
J.Mol.Evol., 89, 2021
|
|
5F5W
 
 | |
6D4K
 
 | |
6D51
 
 | | Crystal structure of L,D-transpeptidase 3 from Mycobacterium tuberculosis in complex with a faropenem-derived adduct | | Descriptor: | ACETYL GROUP, CALCIUM ION, Probable L,D-transpeptidase 3 | | Authors: | Libreros, G.A, Dias, M.V.B. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for the Interaction and Processing of beta-Lactam Antibiotics by l,d-Transpeptidase 3 (LdtMt3) from Mycobacterium tuberculosis.
ACS Infect Dis, 5, 2019
|
|
4JGX
 
 | | The Structure of Sterol Carrier Protein 2 from the Yeast Yarrowia Lipolytica | | Descriptor: | CITRIC ACID, Fatty acid-binding protein, PALMITIC ACID | | Authors: | De Berti, F.P, Capaldi, S, Acierno, J.P, Klinke, S, Monaco, H.L, Ermacora, M.R. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of sterol carrier protein 2 from Yarrowia lipolytica and the evolutionary conservation of a large, non-specific lipid-binding cavity.
J.Struct.Funct.Genom., 14, 2013
|
|
