8A0M
 
 | | Capsular polysaccharide synthesis multienzyme in complex with capsular polymer fragment | | Descriptor: | Bcs3, MAGNESIUM ION, beta-D-ribosyl-(1->1)-D-ribitol-5-phosphate | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
5G5W
 
 | | Structure guided design and discovery of Indazole ethers as highly potent, non-steroidal Glucocorticoid receptor modulators | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-trifluoro-N-[(1R,2S)-1-{[1-(4-fluorophenyl)-1H-indazol-5-yl]oxy}-1-phenylpropan-2-yl]acetamide, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Hemmerling, M, Edman, K, Lepisto, M, Eriksson, A, Ivanova, S, Dahmen, J, Rehwinkel, H, Berger, M, Hendrickx, R, Dearman, M, Jellesmark-Jensen, T, Wissler, L, Hansson, T. | | Deposit date: | 2016-06-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Indazole Ethers as Novel, Potent, Non-Steroidal Glucocorticoid Receptor Modulators.
Bioorg.Med.Chem.Lett., 26, 2017
|
|
7NTN
 
 | | The structure of RRM domain of human TRMT2A at 2 A resolution | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Davydova, E, Janowski, R, Witzenberger, M, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4I5M
 
 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4EWI
 
 | | Crystal structure of the NLRP4 Pyrin domain | | Descriptor: | CHLORIDE ION, NACHT, LRR and PYD domains-containing protein 4, ... | | Authors: | Eibl, C, Hessenberger, M, Puehringer, S, Page, R, Diederichs, K, Peti, W. | | Deposit date: | 2012-04-27 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and Functional Analysis of the NLRP4 Pyrin Domain.
Biochemistry, 51, 2012
|
|
8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
2JUY
 
 | | NMR ensemble of Neopetrosiamide A | | Descriptor: | Neopetrosiamide A | | Authors: | Austin, P, Williams, D.E, Heller, M, McIntosh, L.P, Andersen, R.J, Roberge, M, Roskelley, C.D. | | Deposit date: | 2007-09-05 | | Release date: | 2008-08-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR ensemble of Neopetrosiamide A
To be Published
|
|
2CH5
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
2CH6
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with ADP and glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-ACETYL-D-GLUCOSAMINE KINASE, alpha-D-glucopyranose | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
4H1V
 
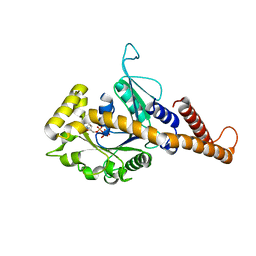 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | Descriptor: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
4N1K
 
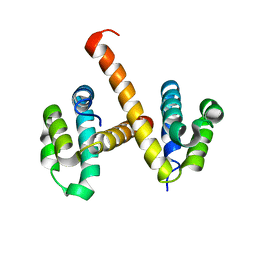 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1J
 
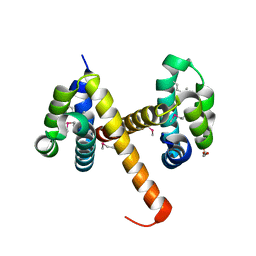 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1L
 
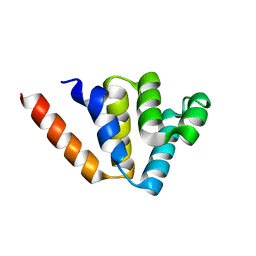 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2NBT
 
 | | NEURONAL BUNGAROTOXIN, NMR, 10 STRUCTURES | | Descriptor: | NEURONAL BUNGAROTOXIN | | Authors: | Oswald, R.E, Sutcliffe, M.J, Bamberger, M, Loring, R.H, Braswell, E, Dobson, C.M. | | Deposit date: | 1997-10-29 | | Release date: | 1998-03-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: calculation of tertiary structure using systematic homologous model building, dynamical simulated annealing, and restrained molecular dynamics.
Biochemistry, 31, 1992
|
|
8ZRA
 
 | |
6F9A
 
 | | Solution structure of the MRH domain of Yos9 complexed with alpha3,alpha6-Man5 | | Descriptor: | ER quality-control lectin | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MRH domain of Yos9 complexed with alpha3,alpha6-Man5
To Be Published
|
|
6F99
 
 | |
3PK2
 
 | | Artificial Transfer Hydrogenases for the Enantioselective Reduction of Cyclic Imines | | Descriptor: | IRIDIUM (III) ION, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Schirmer, T, Heinisch, T. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Artificial transfer hydrogenases for the enantioselective reduction of cyclic imines.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
5LKQ
 
 | | Protease domain of RadA | | Descriptor: | DNA repair protein RadA | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
5LKM
 
 | | RadA bound to dTDP | | Descriptor: | DNA repair protein RadA, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Rapisarda, C, Remaut, H, Fornzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
2MJK
 
 | |
4CSV
 
 | | Tyrosine kinase AS - a common ancestor of Src and Abl bound to Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, SRC-ABL TYROSINE KINASE ANCESTOR | | Authors: | Kutter, S, Wilson, C, Cruz, L, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Kern, D. | | Deposit date: | 2014-03-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
