5Q0W
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2YJH
 
 | | Thiol Peroxidase from Yersinia Psuedotuberculosis, inactive mutant C61S | | Descriptor: | THIOL PEROXIDASE | | Authors: | Beckham, K.S.H, Gabrielsen, M, Wang, D, Roe, A.J. | | Deposit date: | 2011-05-19 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterisation of Tpx from Yersinia Pseudotuberculosis Reveals Insights Into the Binding of Salicylidene Acylhydrazide Compounds.
Plos One, 7, 2012
|
|
2XPD
 
 | | Reduced Thiol peroxidase (Tpx) from yersinia Pseudotuberculosis | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, THIOL PEROXIDASE | | Authors: | Gabrielsen, M, Zetterstrom, C.E, Wang, D, Elofsson, M, Roe, A.J. | | Deposit date: | 2010-08-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterisation of Tpx from Yersinia Pseudotuberculosis Reveals Insights Into the Binding of Salicylidene Acylhydrazide Compounds.
Plos One, 7, 2012
|
|
2XPE
 
 | | Oxidised Thiol peroxidase (Tpx) from Yersinia pseudotuberculosis | | Descriptor: | THIOL PEROXIDASE | | Authors: | Gabrielsen, M, Zetterstrom, C.E, Wang, D, Elofsson, M, Roe, A.J. | | Deposit date: | 2010-08-26 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterisation of Tpx from Yersinia Pseudotuberculosis Reveals Insights Into the Binding of Salicylidene Acylhydrazide Compounds.
Plos One, 7, 2012
|
|
5HZP
 
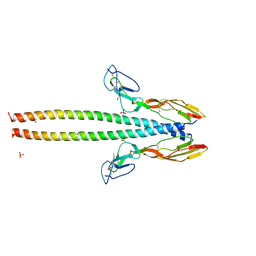 | | Structure of human C4b-binding protein alpha chain CCP domains 1 and 2 in complex with the hypervariable region of group A Streptococcus M49 protein. | | Descriptor: | C4b-binding protein alpha chain, M protein, serotype 49, ... | | Authors: | Buffalo, C.Z, Bahn-Suh, A.J, Ghosh, P. | | Deposit date: | 2016-02-02 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein.
Nat Microbiol, 1, 2016
|
|
5HYP
 
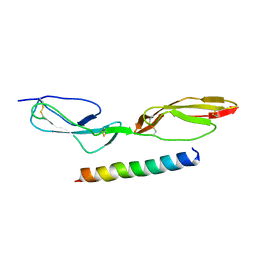 | |
5HYT
 
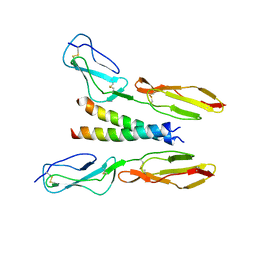 | |
5HZD
 
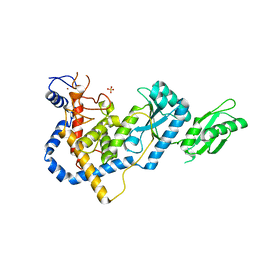 | | RNA Editing TUTase 1 from Trypanosoma brucei | | Descriptor: | 3' terminal uridylyl transferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5HYU
 
 | |
5I49
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP analog UMPNPP | | Descriptor: | 3' terminal uridylyl transferase, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5I0Q
 
 | | Structure of human C4b-binding protein alpha chain CCP domains 1 and 2 in complex with the hypervariable region of mutant group A Streptococcus M2 (K65A, N66A) protein | | Descriptor: | C4b-binding protein alpha chain, M protein, serotype 2.1 | | Authors: | Buffalo, C.Z, Bahn-Suh, A.J, Ghosh, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-20 | | Last modified: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein.
Nat Microbiol, 1, 2016
|
|
5IDO
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP | | Descriptor: | 3' terminal uridylyl transferase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
7KM6
 
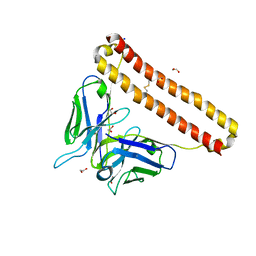 | | APOBEC3B antibody 5G7 Fv-clasp | | Descriptor: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
7KPY
 
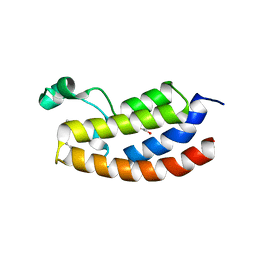 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7JX0
 
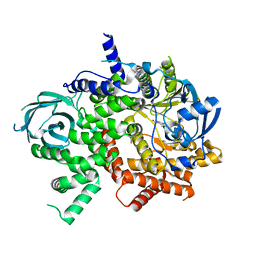 | | NVS-PI3-4 bound to the PI3Kg catalytic subunit p110 gamma | | Descriptor: | N~3~-{[5-(4-acetylphenyl)-4-methyl-1,3-thiazol-2-yl]carbamoyl}-N-tert-butyl-beta-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Rathinaswamy, M.K, Harris, N.J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Disease related mutations in PI3K gamma disrupt regulatory C-terminal dynamics and reveal a path to selective inhibitors.
Elife, 10, 2021
|
|
7JWZ
 
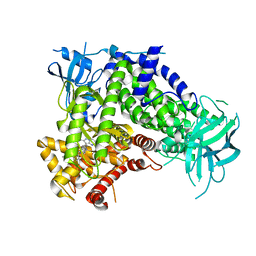 | | IPI-549 bound to the PI3Kg catalytic subunit p110 gamma | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Rathinaswamy, M.K, Harris, N.J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Disease related mutations in PI3K gamma disrupt regulatory C-terminal dynamics and reveal a path to selective inhibitors.
Elife, 10, 2021
|
|
7JWE
 
 | |
5Q0L
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{2-[4-(hydroxymethyl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Y
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[6-(1H-pyrazol-1-yl)pyridin-3-yl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1F
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5-fluoro-1H-benzimidazol-1-yl]acetyl}amino)cyclohexane-1-carboxylic acid | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0S
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(2-cyanophenyl)-2-cyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
