2V5D
 
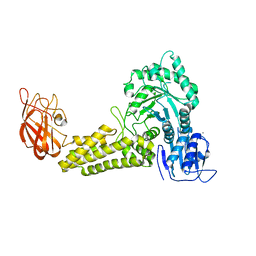 | | Structure of a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. | | Descriptor: | CALCIUM ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
2A2R
 
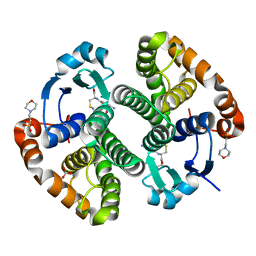 | | Crystal Structure of Glutathione Transferase Pi in complex with S-nitrosoglutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
2A2S
 
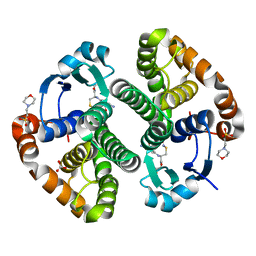 | | Crystal Structure of Human Glutathione Transferase in complex with S-nitrosoglutathione in the absence of reducing agent | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-[1-(CARBOXYLATOMETHYLCARBAMOYL)-2-NITROSOSULFANYL-ETHYL]AMINO-5-OXO-PENTANOATE, CALCIUM ION, ... | | Authors: | Parker, L.J, Morton, C.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1
Protein Sci., 15, 2006
|
|
1ZGN
 
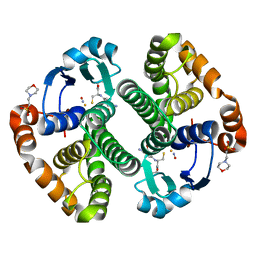 | | Crystal Structure of the Glutathione Transferase Pi in Complex with Dinitrosyl-diglutathionyl Iron Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE (III) ION, GLUTATHIONE, ... | | Authors: | Parker, L.J, Adams, J.J, Parker, M.W. | | Deposit date: | 2005-04-21 | | Release date: | 2005-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrosylation of human glutathione transferase P1-1 with dinitrosyl diglutathionyl iron complex in vitro and in vivo
J.Biol.Chem., 280, 2005
|
|
8VVM
 
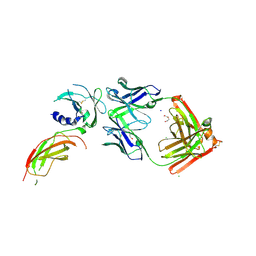 | | Structure of FabS1CE1-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VU1
 
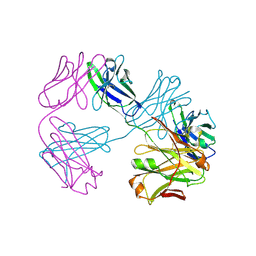 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (trigonal form) | | Descriptor: | S1CE3 VARIANT OF FAB-EPR-1 heavy chain, S1CE3 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VVO
 
 | | Structure of FabS1CE2-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | CHLORIDE ION, Erythropoietin receptor, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VUA
 
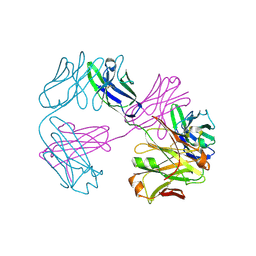 | | Structure of FabS1CE1-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | S1CE1 VARIANT OF FAB-EPR-1 heavy chain, S1CE1 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VTR
 
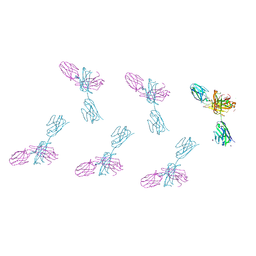 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VU4
 
 | | Structure of FabS1CE4-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VTP
 
 | | Structure of FabS1CE-EPR-1, a high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VUI
 
 | | Structure of FabS1CE-EPR-1, an elbow-locked Fab, in complex with the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VUC
 
 | | Structure of FabS1CE2-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8TRT
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TS5
 
 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7I
 
 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7G
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRS
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T9B
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) in complex with the C-terminal FN3 domain of EphA2 | | Descriptor: | CK variant of Fab F1 heavy chain, CK variant of Fab F1 light chain, Ephrin type-A receptor 2 | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Engineered Antigen-binding Fragments for Enhanced Crystallization of Antibody:Antigen Complexes
To be Published
|
|
3CWM
 
 | | Crystal structure of alpha-1-antitrypsin complexed with citrate | | Descriptor: | Alpha-1-antitrypsin, CITRIC ACID | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
3CWL
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
2JNK
 
 | |
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2O4E
 
 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
