5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
8QQC
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra single crystal | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | VMXm - A sub-micron focus macromolecular crystallography beamline at Diamond Light Source.
J.Synchrotron Radiat., 31, 2024
|
|
3FXU
 
 | | Crystal structure of TsaR in complex with its effector p-toluenesulfonate | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
2NQI
 
 | | Calpain 1 proteolytic core inactivated by WR13(R,R), an epoxysuccinyl-type inhibitor. | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N~2~-[(2S)-2-{[(2R)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]AMINO}PENT-4-ENOYL]-L-ARGINYL-L-TRYPTOPHANAMIDE | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
6TIL
 
 | |
5NQU
 
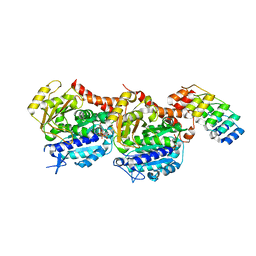 | | Tubulin Darpin cryo structure | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
6TM2
 
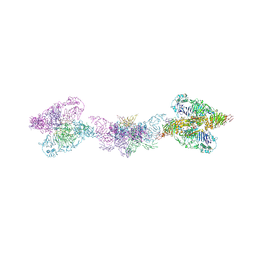 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
6NTS
 
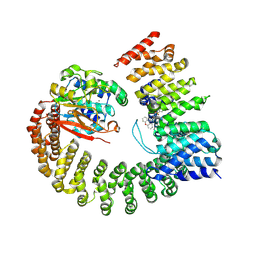 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
2MPW
 
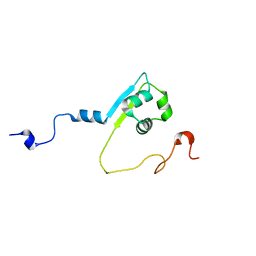 | | Solution structure of the LysM region of the E. coli Intimin periplasmic domain | | Descriptor: | Intimin | | Authors: | Coles, M, Chaubey, M, Leo, J.C, Linke, D, Schuetz, M.C, Goetz, F, Autenrieth, I.B. | | Deposit date: | 2014-06-05 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan.
Mol.Microbiol., 95, 2015
|
|
6YS5
 
 | | Acinetobacter baumannii ribosome-amikacin complex - 30S subunit head | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
5NM5
 
 | | Tubulin Darpin room-temperature structure in complex with Colchicine determined by serial millisecond crystallography | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
6NJJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN14770 | | Descriptor: | (4-{[2-(3-chlorophenyl)-6-(trifluoromethyl)pyridin-4-yl]methyl}phenyl)acetic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
1IGB
 
 | | AEROMONAS PROTEOLYTICA AMINOPEPTIDASE COMPLEXED WITH THE INHIBITOR PARA-IODO-D-PHENYLALANINE HYDROXAMATE | | Descriptor: | AMINOPEPTIDASE, PARA-IODO-D-PHENYLALANINE HYDROXAMIC ACID, ZINC ION | | Authors: | Chevrier, B, D'Orchymont, H, Schalk, C, Tarnus, C, Moras, D. | | Deposit date: | 1996-02-27 | | Release date: | 1996-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit.
Eur.J.Biochem., 237, 1996
|
|
6Y0P
 
 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | Descriptor: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
4YN0
 
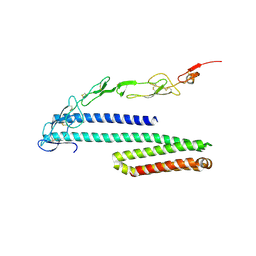 | | Crystal structure of APP E2 domain in complex with DR6 CRD domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amyloid beta A4 protein, MAGNESIUM ION, ... | | Authors: | Xu, K, Nikolov, D. | | Deposit date: | 2015-03-08 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of DR6 in complex with the amyloid precursor protein provides insight into death receptor activation.
Genes Dev., 29, 2015
|
|
6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
1LNE
 
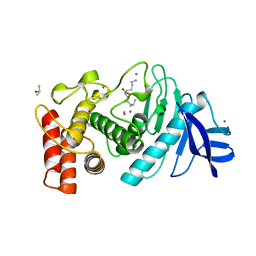 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CADMIUM ION, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
6FK6
 
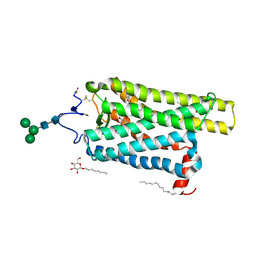 | | Crystal structure of N2C/D282C stabilized opsin bound to RS01 | | Descriptor: | (2~{S})-2-(4-chlorophenyl)-3-methyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4IXQ
 
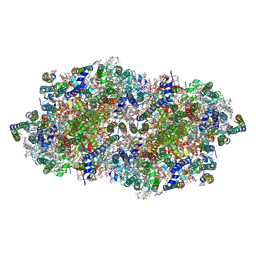 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
7MJR
 
 | | Vip4Da2 toxin structure | | Descriptor: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | Authors: | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
7A1B
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6-dibromo-1H-triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6FKD
 
 | | Crystal structure of N2C/D282C stabilized opsin bound to RS16 | | Descriptor: | 5-chloranyl-2-(2-oxidanylidene-2-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-ethyl)-3~{H}-pyridin-6-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK8
 
 | | Crystal structure of N2C/D282C stabilized opsin bound to RS08 | | Descriptor: | (2~{R},3~{S})-3-azanyl-2-(4-chlorophenyl)-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
