8OYB
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (30 microsecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY4
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (300 ps pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
6FIX
 
 | | antitoxin GraA in complex with its operator | | Descriptor: | DNA (30-MER), XRE family transcriptional regulator | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
3BSQ
 
 | | Crystal structure of human kallikrein 7 produced as a secretion protein in E.coli | | Descriptor: | Kallikrein-7, SULFATE ION | | Authors: | Fernandez, I.S, Standker, L, Magert, H.J, Forssmann, W.G, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human epidermal kallikrein 7 (hK7) synthesized directly in its native state in E. coli: insights into the atomic basis of its inhibition by LEKTI domain 6 (LD6)
J.Mol.Biol., 377, 2008
|
|
2KLC
 
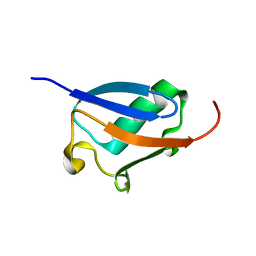 | | NMR solution structure of human ubiquitin-like domain of ubiquilin 1, Northeast Structural Genomics Consortium (NESG) target HT5A | | Descriptor: | Ubiquilin-1 | | Authors: | Doherty, R.S, Dhe-Paganon, S, Fares, C, Lemak, S, Gutmanas, A, Garcia, M, Yee, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ubiquitin-like domain of ubiquilin 1
To be Published
|
|
2KP6
 
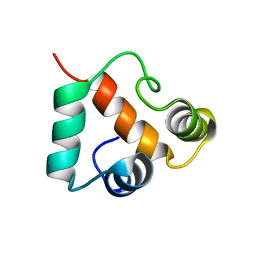 | | Solution NMR structure of protein CV0237 from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESG) target CvT1 | | Descriptor: | Uncharacterized protein | | Authors: | Fares, C, Lemak, A, Yee, A, Garcia, M, Montelione, G.T, Arrowsmith, C.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein CV0237 from Chromobacterium violaceum
TO BE PUBLISHED
|
|
8P9Y
 
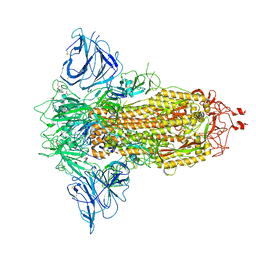 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
8P99
 
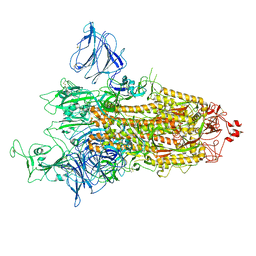 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
4LR6
 
 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4M3B
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
7SBD
 
 | |
4MVL
 
 | |
7SSE
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | Authors: | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
9FEF
 
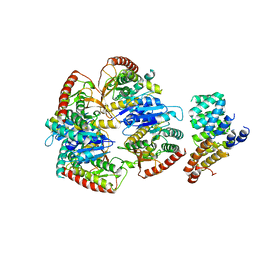 | | Cryo-EM structure of Trypanosoma cruzi (MDH)4-PEX5 complex | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Lipinski, O, Sonani, R.R, Blat, A, Jemiola-Rzeminska, M, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2024-05-19 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of Trypanosoma cruzi (MDH)4-PEX5 complex
To Be Published
|
|
9FGR
 
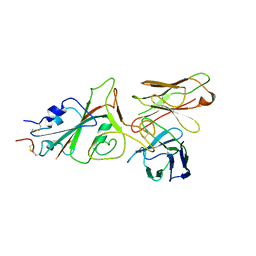 | | SARS-CoV-2 (wuhan variant) Spike protein in complex with the single chain fragment scFv76-77 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, scFv76-77 single chain fragment | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of scFv76-77 in complex with SARS-CoV-2 Spike protein
To Be Published
|
|
9FGS
 
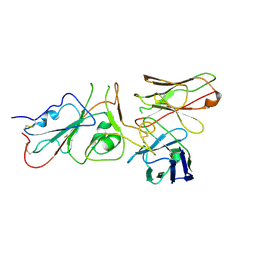 | | SARS-CoV-2 (wuhan variant) Spike protein in complex with the single chain fragment scFv41N (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single Chain fragment scFv41N, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of scFv41N in complex with SARS-CoV-2 Spike protein
To Be Published
|
|
9G0B
 
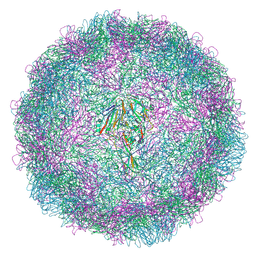 | |
7AKX
 
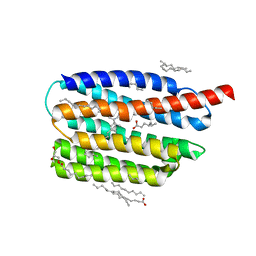 | | Crystal structure of the viral rhodopsin OLPVR1 in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
9F7W
 
 | |
9FVO
 
 | | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Endothiapepsin, ... | | Authors: | Falke, S, Senst, J.M, Guenther, S, Meents, A. | | Deposit date: | 2024-06-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin
To Be Published
|
|
8Y85
 
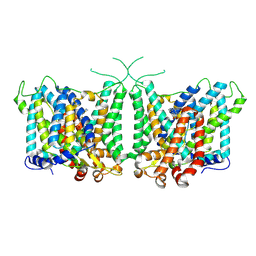 | | Human AE3 with NaHCO3- and DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
5AZ1
 
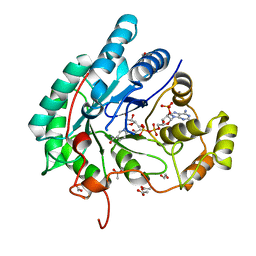 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
8XZ3
 
 | |
9FEE
 
 | | Cryo-EM structure of Trypanosoma cruzi glycosomal malate dehydrogenase | | Descriptor: | malate dehydrogenase | | Authors: | Lipinski, O, Sonani, R.R, Blat, A, Jemiola-Rzeminska, M, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2024-05-19 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structure of Trypanosoma cruzi glycosomal malate dehydrogenase
To Be Published
|
|
