6IBV
 
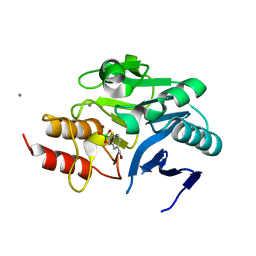 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
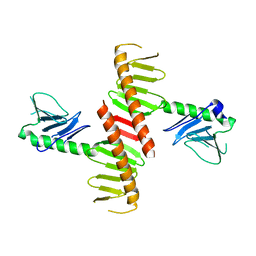
6N45
 
 | |
5EGM
 
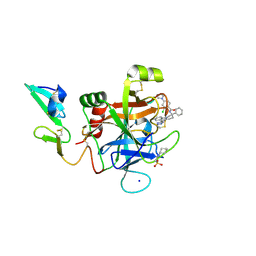 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2P7N
 
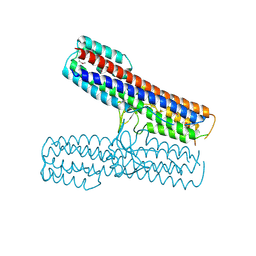 | | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69. | | Descriptor: | Pathogenicity island 1 effector protein | | Authors: | Benach, J, Abashidze, M, Seetharaman, J, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69.
To be Published
|
|
2OTG
 
 | | Rigor-like structures of muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2007-02-08 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Rigor-like Structures from Muscle Myosins Reveal Key Mechanical Elements in the Transduction Pathways of This Allosteric Motor.
Structure, 15, 2007
|
|
3OLD
 
 | | Crystal structure of alpha-amylase in complex with acarviostatin I03 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Qin, X, Ren, L. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes
J.Struct.Biol., 174, 2011
|
|
6JFM
 
 | | Mitofusin2 (MFN2)_T111D | | Descriptor: | ACETATE ION, CALCIUM ION, Mitofusin-2,Mitofusin-2 | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
2P6Y
 
 | | X-ray structure of the protein Q9KM02_VIBCH from Vibrio cholerae at the resolution 1.63 A. Northeast Structural Genomics Consortium target VcR80. | | Descriptor: | Hypothetical protein VCA0587, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Chen, C.X, Wang, C, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-19 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure of the protein Q9KM02_VIBCH from Vibrio cholerae at the resolution 1.63 A.
To be Published
|
|
1SVY
 
 | | SEVERIN DOMAIN 2, 1.75 ANGSTROM CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, SEVERIN, SODIUM ION | | Authors: | Puius, Y.A, Fedorov, E.V, Eichinger, L, Sullivan, M, Schleicher, M, Almo, S.C. | | Deposit date: | 1998-08-10 | | Release date: | 1999-08-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the functional surface of domain 2 in the gelsolin superfamily.
Biochemistry, 39, 2000
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
2Q00
 
 | | Crystal structure of the P95883_SULSO protein from Sulfolobus solfataricus. NESG target SsR10. | | Descriptor: | Orf c02003 protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Owens, L, Ma, L.-C, Cunningham, K, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the P95883_SULSO protein from Sulfolobus solfataricus.
To be Published
|
|
2PHR
 
 | | Pterocarpus angolensis lectin (PAL) in complex with Man-7D1 | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Garcia-Pino, A, Buts, L, Wyns, L, Imberty, A, Loris, R. | | Deposit date: | 2007-04-12 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How a plant lectin recognizes high mannose oligosaccharides
Plant Physiol., 144, 2007
|
|
8IEJ
 
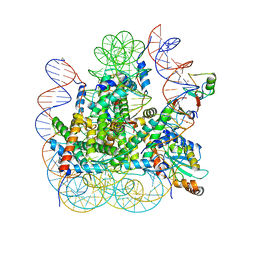 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
2Q88
 
 | | Crystal structure of EhuB in complex with ectoine | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
4L29
 
 | | Structure of wtMHC class I with NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-04 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
8I1O
 
 | |
8I1M
 
 | | Crystal structure of oxidated APSK1 domain from human PAPSS1 in complex with APS and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, PAPSS1 protein, ... | | Authors: | Zhang, L, Song, W.Y, Zhang, L. | | Deposit date: | 2023-01-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Redox switching mechanism of the adenosine 5'-phosphosulfate kinase domain (APSK2) of human PAPS synthase 2.
Structure, 31, 2023
|
|
8I1N
 
 | |
6H0B
 
 | | Crystal structure of the human GalNAc-T4 in complex with UDP, manganese and the diglycopeptide 6. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, ALA-THR-GLY-ALA-GLY-ALA-GLY-ALA-GLY-THR-THR-PRO-GLY-PRO-GLY, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Coelho, H, Lira-Navarrete, E, Raich, L, Companon, I, Diniz, A, Lagartera, L, Jimenez-Barbero, J, Clausen, H, Rovira, C, Marcelo, F, Corzana, F, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2018-07-08 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights into the Catalytic-Domain-Mediated Short-Range Glycosylation Preferences of GalNAc-T4.
ACS Cent Sci, 4, 2018
|
|
7WEC
 
 | |
7WEA
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEB
 
 | |
7WEE
 
 | |
