3PVD
 
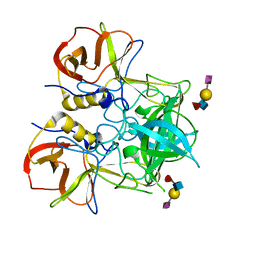 | | Crystal structure of P domain dimer of Norovirus VA207 complexed with 3'-sialyl-Lewis x tetrasaccharide | | Descriptor: | Capsid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3PUN
 
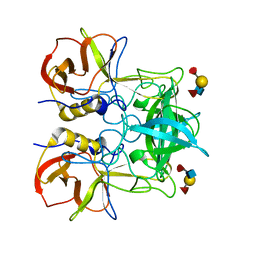 | | Crystal structure of P domain dimer of Norovirus VA207 with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
1WDF
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
8HJT
 
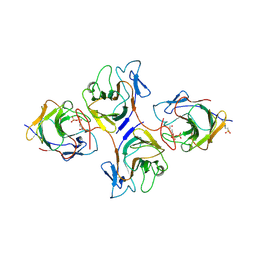 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6UOX
 
 | | Structure of itraconazole-bound NPC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-bromo-4-{4-[4-({(2R,4S)-2-(2,4-dichlorophenyl)-2-[(1H-1,2,4-triazol-1-yl)methyl]-1,3-dioxolan-4-yl}methoxy)phenyl]piperazin-1-yl}phenyl)-2-[(2S)-butan-2-yl]-2,4-dihydro-3H-1,2,4-triazol-3-one, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis for itraconazole-mediated NPC1 inhibition.
Nat Commun, 11, 2020
|
|
1WDG
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
9MS8
 
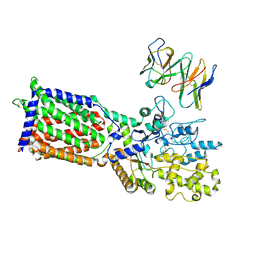 | | PTCH1 in complex with Fab6H3 | | Descriptor: | 6H3 Fab heavy chain, 6H3 Fab light chain, Protein patched homolog 1 | | Authors: | Hu, Q, Qi, X, Li, X. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Inhibiting hedgehog signal by a patched-1 antibody.
Cell Discov, 11, 2025
|
|
3V32
 
 | | Crystal structure of MCPIP1 N-terminal conserved domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V34
 
 | | Crystal structure of MCPIP1 conserved domain with magnesium ion in the catalytic center | | Descriptor: | MAGNESIUM ION, Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
4F8E
 
 | | Crystal structure of human PRS1 D52H mutant | | Descriptor: | MAGNESIUM ION, Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A small disturbance, but a serious disease: the possible mechanism of D52H-mutant of human PRS1 that causes gout
Iubmb Life, 65, 2013
|
|
3QXL
 
 | | Crystal structure of the CDC25 Domain from Ral-specific Guanine-nucleotide Exchange Factor RalGPS1a | | Descriptor: | Ras-specific guanine nucleotide-releasing factor RalGPS1 | | Authors: | Peng, W, Xu, J, Guan, X, Sun, Y, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural study of the Cdc25 domain from Ral-specific guanine-nucleotide exchange factor RalGPS1a.
Protein Cell, 2, 2011
|
|
3V33
 
 | | Crystal structure of MCPIP1 conserved domain with zinc-finger motif | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3PUM
 
 | | Crystal structure of P domain dimer of Norovirus VA207 | | Descriptor: | Capsid | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
4MZD
 
 | | High resolution crystal structure of the nisin leader peptidase NisP from Lactococcus lactis | | Descriptor: | Nisin leader peptide-processing serine protease NisP | | Authors: | Rao, Z.H, Xu, Y.Y, Li, X, Yang, W. | | Deposit date: | 2013-09-30 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the nisin leader peptidase NisP revealing a C-terminal autocleavage activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QWQ
 
 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | Descriptor: | Response regulator SaeR | | Authors: | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | Deposit date: | 2014-07-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RDL
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | Descriptor: | Capsid | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
6LTP
 
 | | Crystal structure of Cas12i2 binary complex | | Descriptor: | Cas12i2, crRNA (56-mer RNA) | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6OT0
 
 | | Structure of human Smoothened-Gi complex | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Fab heavy chain, Fab light chain, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi.
Nature, 571, 2019
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
1QST
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QSR
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QXK
 
 | | Monoacid-Based, Cell Permeable, Selective Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | 2-{4-[2-ACETYLAMINO-3-(4-CARBOXYMETHOXY-3-HYDROXY-PHENYL)-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZOIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Liu, G, Abad-Zapatero, C, Pei, Z, Szczepankiewick, B.G, Li, X, Zhang, T, Hutchins, C.W, Hajduk, P.J, Ballaron, S.J, Stashko, M.A, Lubben, T.H, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Monoacid-Based, Cell Permeable, Selective
Inhibitor of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
1QSN
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | Descriptor: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-22 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
6IY3
 
 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
