7LYI
 
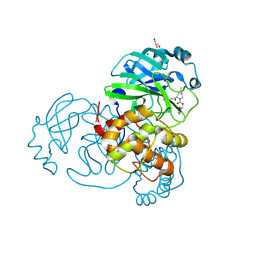 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
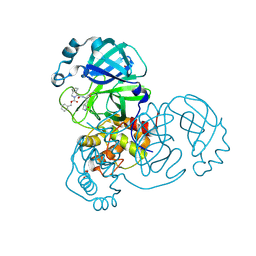 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
4U93
 
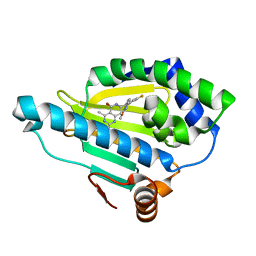 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4W7T
 
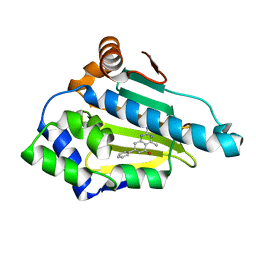 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
5TRL
 
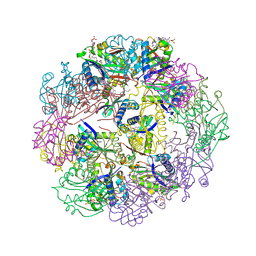 | |
4ZS3
 
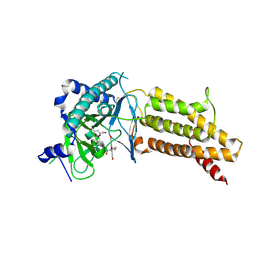 | | Structural complex of 5-aminofluorescein bound to the FTO protein | | Descriptor: | 2-OXOGLUTARIC ACID, 5-amino-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
4ZS2
 
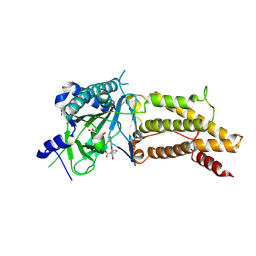 | | Structural complex of FTO/fluorescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
5TRM
 
 | |
5B64
 
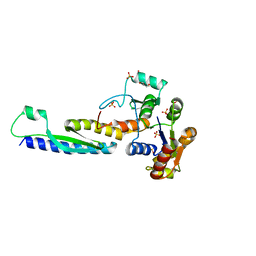 | | A novel binding mode of MAGUK GK domain revealed by DLG GK domain in complex with KIF13B MBS domain | | Descriptor: | DLG GK, GLYCEROL, Protein Kif13b, ... | | Authors: | Shang, Y, Zhu, J, Zhang, M. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An Atypical MAGUK GK Target Recognition Mode Revealed by the Interaction between DLG and KIF13B
Structure, 24, 2016
|
|
2AIU
 
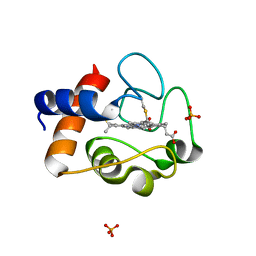 | | Crystal Structure of Mouse Testicular Cytochrome C at 1.6 Angstrom | | Descriptor: | Cytochrome c, testis-specific, PHOSPHATE ION, ... | | Authors: | Liu, Z, Ye, S, Lin, H, Rao, Z, Liu, X.J. | | Deposit date: | 2005-08-01 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Remarkably high activities of testicular cytochrome c in destroying reactive oxygen species and in triggering apoptosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4YAQ
 
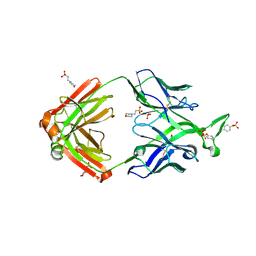 | | Crystal structure of a computationally optimized PG9 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PG9_N100FY Fab heavy chain, ... | | Authors: | Murrell, S, Julien, J.P, Wilson, I.A. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redesigned HIV antibodies exhibit enhanced neutralizing potency and breadth.
J.Clin.Invest., 125, 2015
|
|
5WZE
 
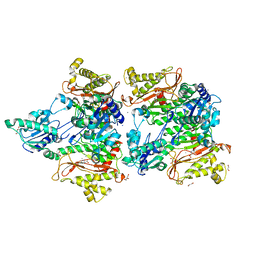 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
8HLT
 
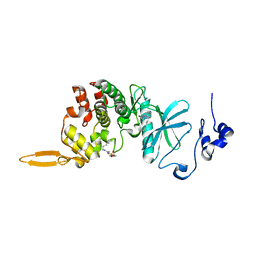 | | The co-crystal structure of DYRK2 with YK-2-99B | | Descriptor: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
7C96
 
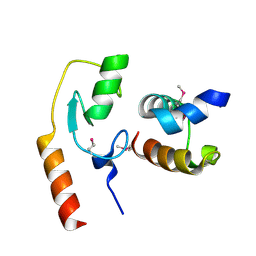 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3K30
 
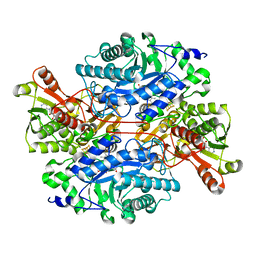 | | Histamine dehydrogenase from Nocardiodes simplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Histamine dehydrogenase, ... | | Authors: | Scott, E.E, Reed, T.M, Limburg, J. | | Deposit date: | 2009-09-30 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histamine dehydrogenase from Nocardioides simplex.
J.Biol.Chem., 285, 2010
|
|
8KAE
 
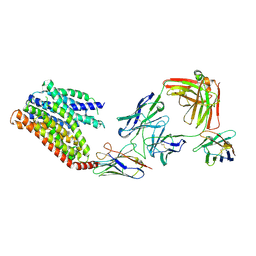 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
4KMA
 
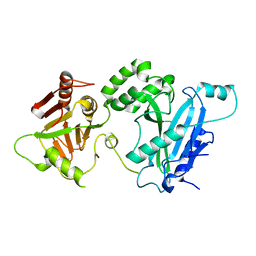 | |
4KMH
 
 | |
4KM9
 
 | |
4KMD
 
 | | Crystal structure of Sufud60-Gli1p | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
4KM8
 
 | | Crystal structure of Sufud60 | | Descriptor: | Suppressor of fused homolog | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
7FD2
 
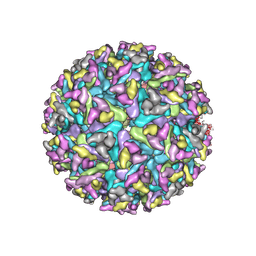 | | Cryo-EM structure of an alphavirus, Getah virus | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, Z, Liu, C, Wang, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of infective Getah virus at 2.8 angstrom resolution determined by cryo-electron microscopy.
Cell Discov, 8, 2022
|
|
