3ACA
 
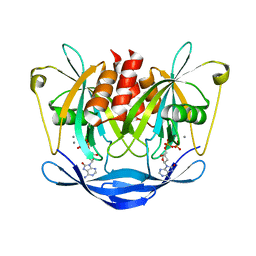 | |
3W3M
 
 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 2 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3WPF
 
 | | Crystal structure of mouse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
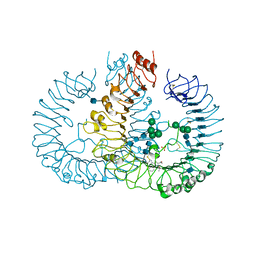
3W3K
 
 | | Crystal structure of human TLR8 in complex with CL075 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-propyl[1,3]thiazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
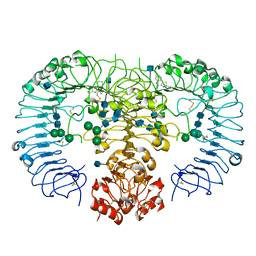
3WPD
 
 | | Crystal structure of horse TLR9 in complex with inhibitory DNA4084 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*GP*GP*AP*TP*GP*GP*G)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3W3J
 
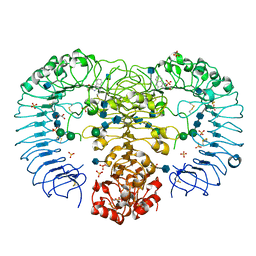 | | Crystal structure of human TLR8 in complex with CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3WPG
 
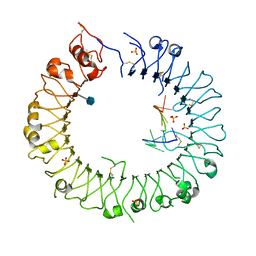 | |
3WPI
 
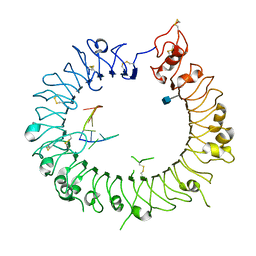 | | Crystal structure of mouse TLR9 in complex with inhibitory DNA_super | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*CP*AP*AP*TP*AP*GP*GP*GP*TP*GP*AP*GP*GP*GP*G)-3'), Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3W3L
 
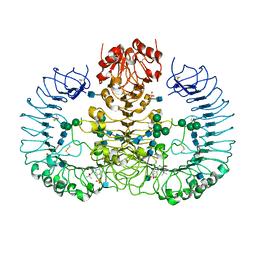 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 1 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3WPE
 
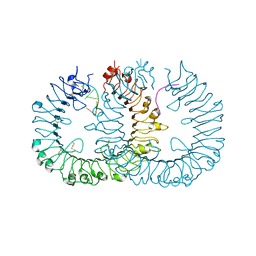 | |
3W3N
 
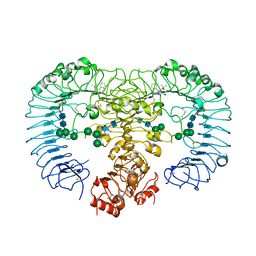 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 3 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3WPB
 
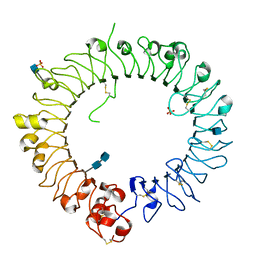 | | Crystal structure of horse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPH
 
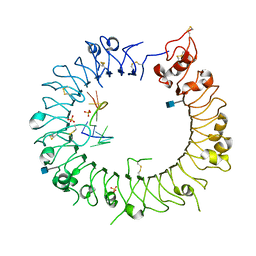 | |
3WPC
 
 | | Crystal structure of horse TLR9 in complex with agonistic DNA1668_12mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*AP*TP*GP*AP*CP*GP*TP*TP*CP*CP*T)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3AC9
 
 | |
1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY5
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE COMPLEX WITH MALEATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY8
 
 | |
2DDQ
 
 | |
1VHR
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE VHR, SULFATE ION | | Authors: | Yuvaniyama, J, Denu, J.M, Dixon, J.E, Saper, M.A. | | Deposit date: | 1996-02-20 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the dual specificity protein phosphatase VHR.
Science, 272, 1996
|
|
3A73
 
 | |
1VDH
 
 | | Structure-based functional identification of a novel heme-binding protein from thermus thermophilus HB8 | | Descriptor: | muconolactone isomerase-like protein | | Authors: | Ebihara, A, Okamoto, A, Kousumi, Y, Yamamoto, H, Masui, R, Ueyama, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
2AY3
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITH 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID | | Descriptor: | 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID, AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1998-08-06 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of Paracoccus denitrificans aromatic amino acid aminotransferase has contrary properties: flexibility and rigidity.
Biochemistry, 38, 1999
|
|
2AY9
 
 | |
2AY6
 
 | |
