6IZ0
 
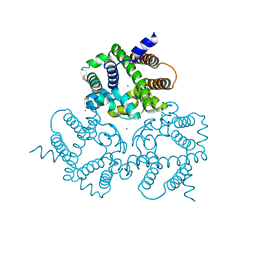 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3UWP
 
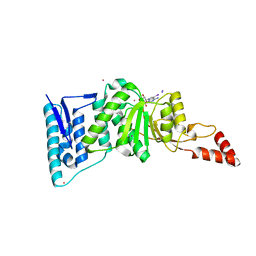 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
2LNW
 
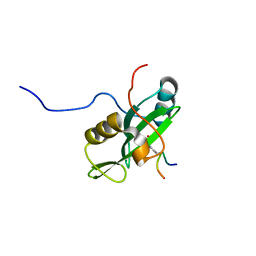 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LNX
 
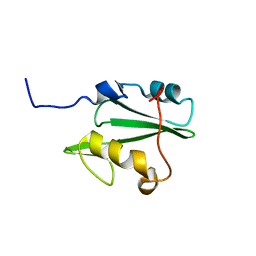 | | Solution structure of Vav2 SH2 domain | | Descriptor: | Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
2LUY
 
 | | Solution structure of the tandem zinc finger domain of fission yeast Stc1 | | Descriptor: | Meiotic chromosome segregation protein P8B7.28c, ZINC ION | | Authors: | He, C, Shi, Y, Bayne, E, Wu, J. | | Deposit date: | 2012-06-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H12
 
 | | The crystal structure of methyltransferase domain of human SET domain-containing protein 2 in complex with S-adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Amaya, M.F, Dong, A, Zeng, H, Mackenzie, F, Bunnage, M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
6OGN
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
6IZ6
 
 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | CALCIUM ION, Trimeric intracellular cation channel type B-B | | Authors: | Liu, X.L, Wang, X.H, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZF
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ3
 
 | |
8JOP
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
5YTK
 
 | | Crystal structure of SIRT3 bound to a leucylated AceCS2 | | Descriptor: | AceCS2-KLeu, LEUCINE, LYSINE, ... | | Authors: | Li, J, Gong, W, Xu, Y. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations
Cell Metab., 27, 2018
|
|
5CPR
 
 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | Descriptor: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Jakob, C.G, Upadhyay, A.K, Sun, C. | | Deposit date: | 2015-07-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
5GTU
 
 | |
5T5G
 
 | | human SETD8 in complex with MS2177 | | Descriptor: | 7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)-N-[5-(pyrrolidin-1-yl)pentyl]quinazolin-4-amine, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
6E68
 
 | | NAMPT co-crystal with inhibitor compound 2 | | Descriptor: | (2E)-N-{4-[1-(3-aminobenzene-1-carbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Waight, A.B, Neumann, C.S. | | Deposit date: | 2018-07-24 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NAMPT co-crystal with inhibitor compound 2
to be published
|
|
6G25
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 4 | | Descriptor: | 3,5-dimethyl-4-(4-pyridin-4-yl-1~{H}-pyrazol-3-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2B
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 8 | | Descriptor: | 4-(3-methyl-5-phenyl-imidazol-4-yl)pyridine, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2O
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound BI-9321 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G29
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 6 | | Descriptor: | 5-methyl-2-piperidin-4-yl-6-pyridin-4-yl-pyridazin-3-one, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2E
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 13 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2F
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 16 | | Descriptor: | 4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G3P
 
 | |
4PHG
 
 | | Crystal structure of Ypt7 covalently modified with GTP | | Descriptor: | GTP-binding protein YPT7, MAGNESIUM ION, N-[3-(propanoylamino)propyl]guanosine 5'-(tetrahydrogen triphosphate), ... | | Authors: | Koch, D, Wiegandt, D, Vieweg, S, Hofmann, F, Wu, Y, Itzen, A, Mueller, M.P, Goody, R.S. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Locking GTPases covalently in their functional states.
Nat Commun, 6, 2015
|
|
