1E0D
 
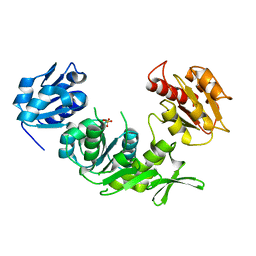 | | UDP-N-Acetylmuramoyl-L-Alanine:D-Glutamate Ligase | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Fanchon, E, Bertrand, J, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-03-24 | | Release date: | 2000-06-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Open" Structures of Murd: Domain Movements and Structural Similarities with Folylpolyglutamate Synthetase.
J.Mol.Biol., 301, 2000
|
|
1CHN
 
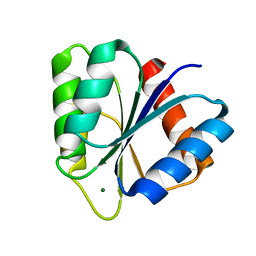 | |
2QK7
 
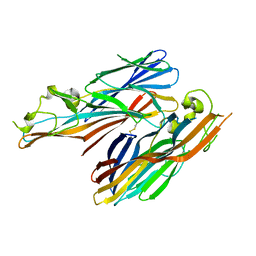 | | A covalent S-F heterodimer of staphylococcal gamma-hemolysin | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Roblin, P, Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2007-07-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of beta-barrel pore-forming toxins.
Proteins, 71, 2008
|
|
1W82
 
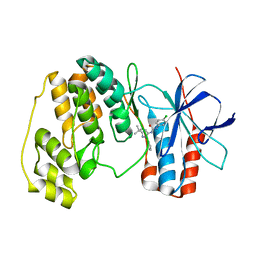 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[(3Z)-5-TERT-BUTYL-2-PHENYL-1,2-DIHYDRO-3H-PYRAZOL-3-YLIDENE]-N'-(4-CHLOROPHENYL)UREA | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
1E6G
 
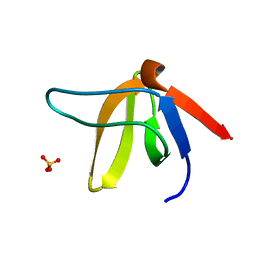 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25I, V53I, V58L MUTANT | | Descriptor: | SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-15 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Strain in the Hydrophobic Core and its Implications for Protein Folding and Design
Nat.Struct.Biol., 9, 2002
|
|
4WZN
 
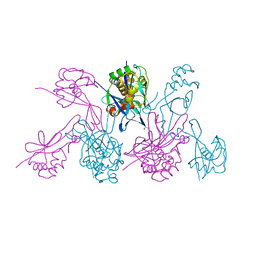 | | CRYSTAL STRUCTURE OF THE 2B PROTEIN SOLUBLE DOMAIN FROM HEPATITIS A VIRUS | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Garriga, D, Vives-Adrian, L, Buxaderas, M, Ferreira-da-Silva, F, Almeida, B, Macedo-Ribeiro, S, Pereira, P.J, Verdaguer, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Host Membrane Remodeling Induced by Protein 2B of Hepatitis A Virus.
J.Virol., 89, 2015
|
|
5ILB
 
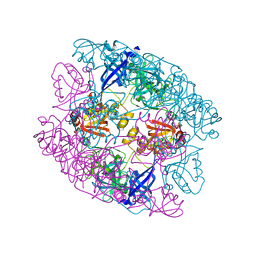 | | Crystal structure of protease domain of Deg2 linked with the PDZ domain of Deg9 | | Descriptor: | Protease Do-like 2, chloroplastic,Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
6S0F
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
2JSD
 
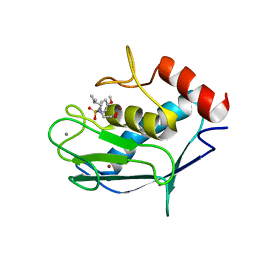 | | Solution structure of MMP20 complexed with NNGH | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-03 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
2JT6
 
 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) | | Descriptor: | 3-[(4'-cyanobiphenyl-4-yl)oxy]-N-hydroxypropanamide, CALCIUM ION, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2IZ9
 
 | | MS2-RNA HAIRPIN (4ONE -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*U ONEP *AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Stonehouse, N.J, Adams, C.J, Fridborg, K, Beigelman, L, Matulic-Adamic, J, Warriner, S.L, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Deletion of a Single Hydrogen Bonding Atom from the MS2 RNA Operator Leads to Dramatic Rearrangements at the RNA-Coat Protein Interface
Nucleic Acids Res., 28, 2000
|
|
3IUC
 
 | | Crystal structure of the human 70kDa heat shock protein 5 (BiP/GRP78) ATPase domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70kDa protein 5 (Glucose-regulated protein, ... | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
5IL9
 
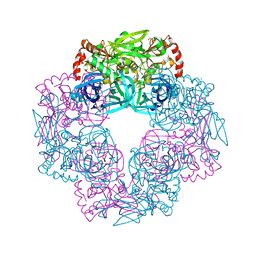 | | Crystal structure of Deg9 | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
2J6R
 
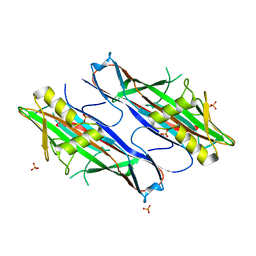 | | FaeG from F4ac ETEC strain GIS26, produced in tobacco plant chloroplast | | Descriptor: | K88 FIMBRIAL PROTEIN, PHOSPHATE ION | | Authors: | Van Molle, I, Joensuu, J.J, Buts, L, Panjikar, S, Kotiaho, M, Bouckaert, J, Wyns, L, Niklander-Teeri, V, De Greve, H. | | Deposit date: | 2006-10-03 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chloroplasts Assemble the Major Subunit Faeg of Escherichia Coli F4 (K88) Fimbriae Into Strand-Swapped Dimers
J.Mol.Biol., 368, 2007
|
|
6SBS
 
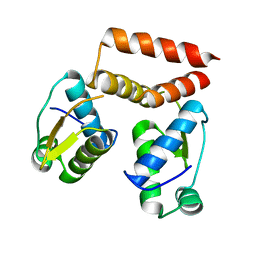 | | YtrA from Sulfolobus acidocaldarius, a GntR-family transcription factor | | Descriptor: | Regulatory protein | | Authors: | Lemmens, L, Valegard, K, Lindas, A.C, Peeters, E, Maes, D. | | Deposit date: | 2019-07-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | YtrASa, a GntR-Family Transcription Factor, Represses Two Genetic Loci Encoding Membrane Proteins inSulfolobus acidocaldarius.
Front Microbiol, 10, 2019
|
|
6IRL
 
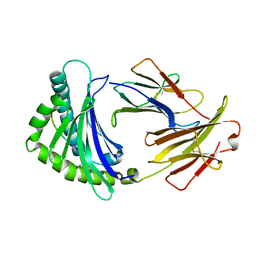 | |
7JU9
 
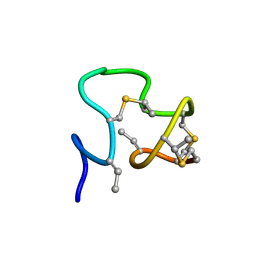 | |
7B7R
 
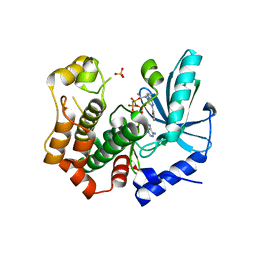 | | MEK1 in complex with compound 4 | | Descriptor: | 2-[5-[ethyl(methyl)amino]imidazo[1,2-a]pyrimidin-7-yl]phenol, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3M
 
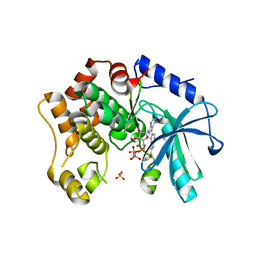 | | MEK1 in complex with compound 6 | | Descriptor: | 1~{H}-indol-2-yl(pyridin-3-yl)methanone, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
4QFC
 
 | |
6SL8
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
3ILN
 
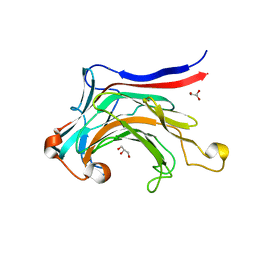 | | X-ray structure of the laminarinase from Rhodothermus marinus | | Descriptor: | CALCIUM ION, GLYCEROL, Laminarinase | | Authors: | Bleicher, L, Golubev, A, Rojas, A.L, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of the thermostability and thermophilicity of laminarinases: X-ray structure of the hyperthermostable laminarinase from Rhodothermus marinus and molecular dynamics simulations.
J.Phys.Chem.B, 115, 2011
|
|
6SK1
 
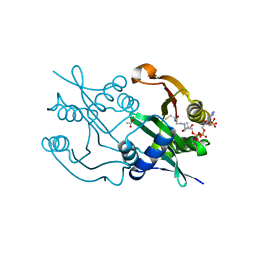 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with coenzyme A | | Descriptor: | ACETATE ION, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
4QFD
 
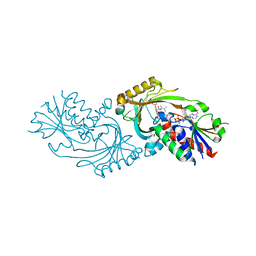 | | Co-crystal structure of compound 2 (3-(7-hydroxy-2-oxo-4-phenyl-2H-chromen-6-yl)propanoic acid) and FAD bound to human DAAO at 2.85A | | Descriptor: | 3-(7-hydroxy-2-oxo-4-phenyl-2H-chromen-6-yl)propanoic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Edwards, T.E, Chun, L, Arakaki, T.L. | | Deposit date: | 2014-05-20 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Novel human D-amino acid oxidase inhibitors stabilize an active-site lid-open conformation.
Biosci.Rep., 34, 2014
|
|
