1SO9
 
 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1JWW
 
 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1KQK
 
 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
2LU5
 
 | | Structure and chemical shifts of Cu(I),Zn(II) superoxide dismutase by solid-state NMR | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Knight, M.J, Pell, A.J, Bertini, I, Felli, I.C, Gonnelli, L, Pierattelli, R, Herrmann, T, Emsley, L, Pintacuda, G. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI6
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
1P6T
 
 | | Structure characterization of the water soluble region of P-type ATPase CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the function of the N-terminal domain of the ATPase CopA from Bacillus subtilis.
J.Biol.Chem., 278, 2003
|
|
1OQ3
 
 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1OQ6
 
 | | solution structure of Copper-S46V CopA from Bacillus subtilis | | Descriptor: | COPPER (II) ION, Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, l, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
2GGP
 
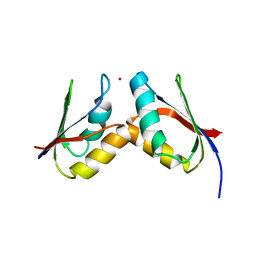 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
2JT5
 
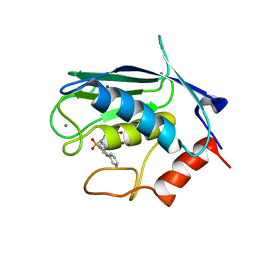 | | solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of n-hydroxy-2-[n-(2-hydroxyethyl)biphenyl-4-sulfonamide] hydroxamic acid (MLC88) | | Descriptor: | CALCIUM ION, N~2~-(biphenyl-4-ylsulfonyl)-N-hydroxy-N~2~-(2-hydroxyethyl)glycinamide, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-19 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JSD
 
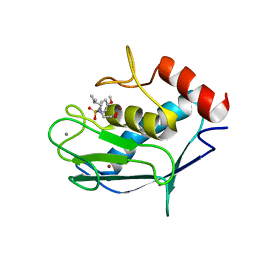 | | Solution structure of MMP20 complexed with NNGH | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-03 | | Release date: | 2007-11-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
2JT6
 
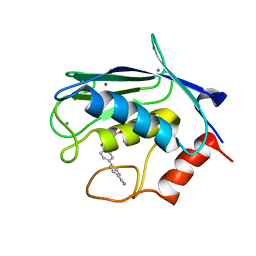 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) | | Descriptor: | 3-[(4'-cyanobiphenyl-4-yl)oxy]-N-hydroxypropanamide, CALCIUM ION, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-23 | | Release date: | 2008-02-19 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JNP
 
 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-3, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
