8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
1JQI
 
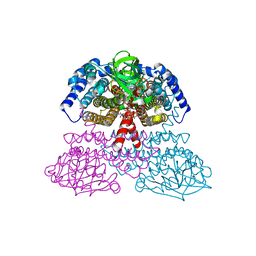 | | Crystal Structure of Rat Short Chain Acyl-CoA Dehydrogenase Complexed With Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, short chain acyl-CoA dehydrogenase | | Authors: | Battaile, K.P, Molin-Case, J, Paschke, R, Wang, M, Bennett, D, Vockley, J, Kim, J.-J.P. | | Deposit date: | 2001-08-07 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J.Biol.Chem., 277, 2002
|
|
1IVH
 
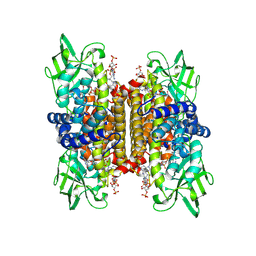 | | STRUCTURE OF HUMAN ISOVALERYL-COA DEHYDROGENASE AT 2.6 ANGSTROMS RESOLUTION: STRUCTURAL BASIS FOR SUBSTRATE SPECIFICITY | | Descriptor: | COENZYME A PERSULFIDE, FLAVIN-ADENINE DINUCLEOTIDE, ISOVALERYL-COA DEHYDROGENASE | | Authors: | Tiffany, K.A, Roberts, D.L, Wang, M, Paschke, R, Mohsen, A.-W.A, Vockley, J, Kim, J.J.P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity,.
Biochemistry, 36, 1997
|
|
5XGQ
 
 | |
5XET
 
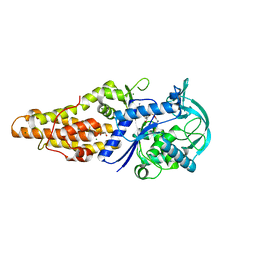 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
2AKJ
 
 | | Structure of spinach nitrite reductase | | Descriptor: | Ferredoxin--nitrite reductase, chloroplast, IRON/SULFUR CLUSTER, ... | | Authors: | Swamy, U, Wang, M, Tripathy, J.N, Kim, S.-K, Hirasawa, M, Knaff, D.B, Allen, J.P. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Biochemistry, 44, 2005
|
|
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4Q5Y
 
 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4IAO
 
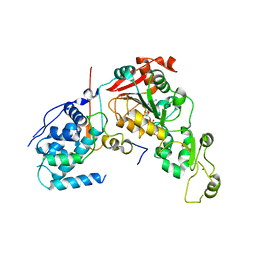 | | Crystal structure of Sir2 C543S mutant in complex with SID domain of Sir4 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent histone deacetylase SIR2, Regulatory protein SIR4, ... | | Authors: | Hsu, H.C, Wang, C.L, Wang, M, Yang, N, Chen, Z, Sternglanz, R, Xu, R.M. | | Deposit date: | 2012-12-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding
Genes Dev., 27, 2013
|
|
5DWK
 
 | | Diacylglycerol Kinase solved by multi crystal multi orientation native SAD | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Weinert, T, Olieric, V, Finke, A.D, Li, D, Caffrey, M, Wang, M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Data-collection strategy for challenging native SAD phasing.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8HBL
 
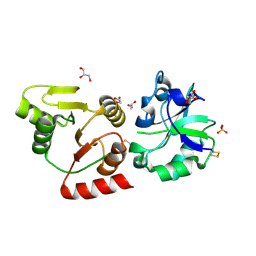 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.58 angstrom resolution) | | Descriptor: | GLYCEROL, LITHIUM ION, Non-structural protein 3, ... | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-10-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
8GQC
 
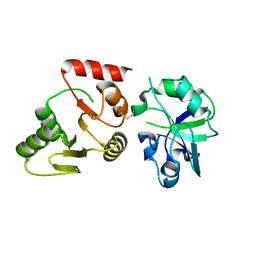 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.35 angstrom resolution) | | Descriptor: | Papain-like protease nsp3 | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
6KY4
 
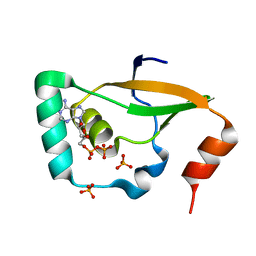 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
3TIB
 
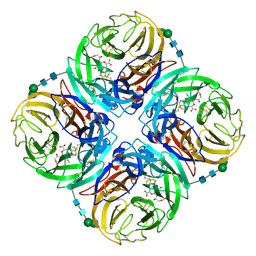 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir octanoate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-9-O-octanoyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI4
 
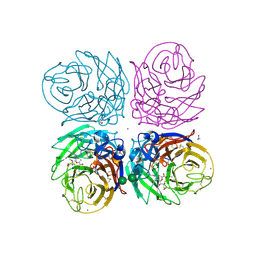 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with laninamivir octanoate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-9-O-octanoyl-D-glycero-D-galacto-non-2-enonic acid, ACETATE ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TIA
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TIC
 
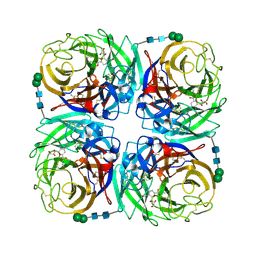 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
1EGE
 
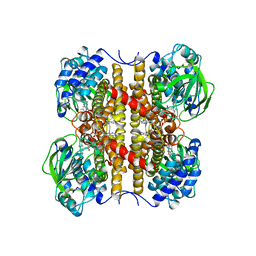 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGD
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGC
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE, OCTANOYL-COENZYME A | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
5ZTE
 
 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | Descriptor: | 2-Cys peroxiredoxin BAS1, chloroplastic | | Authors: | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
