4U0Q
 
 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to basigin | | Descriptor: | Basigin, Reticulocyte binding protein 5 | | Authors: | Wright, K.E, Hjerrild, K.A, Bartlett, J, Douglas, A.D, Jin, J, Brown, R.E, Ashfield, R, Clemmensen, S.B, de Jongh, W.A, Draper, S.J, Higgins, M.K. | | Deposit date: | 2014-07-14 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies.
Nature, 515, 2014
|
|
3NFR
 
 | | Casimiroin analog inhibitor of quinone reductase 2 | | Descriptor: | 6,8-dimethoxy-1,4-dimethylquinolin-2(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Sturdy, M, Mesecar, A.D, Jermihov, K, Cushman, M, Maiti, A. | | Deposit date: | 2010-06-10 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | X-ray Crystallographic Structure Activity Relationship (SAR) of Casimiroin and its Analogs Bound to Human Quinone Reductase 2
To be Published
|
|
4U0R
 
 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to monoclonal antibody 9AD4 | | Descriptor: | Monoclonal antibody 9AD4 heavy chain, Monoclonal antibody 9AD4 light chain, Reticulocyte binding protein 5 | | Authors: | Wright, K.E, Hjerrild, K.A, Bartlett, J, Douglas, A.D, Jin, J, Brown, R.E, Ashfield, R, Clemmensen, S.B, de Jongh, W.A, Draper, S.J, Higgins, M.K. | | Deposit date: | 2014-07-14 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies.
Nature, 515, 2014
|
|
2WYG
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1R)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2WIH
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-848125 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1,4,4-TETRAMETHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
2WZM
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
4U4T
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound inward-open state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
2WEH
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
4U9L
 
 | | Structure of a membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MAGNESIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
4U9N
 
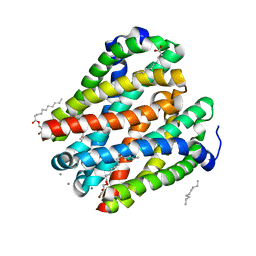 | | Structure of a membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MANGANESE (II) ION, Magnesium transporter MgtE, ... | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
2WEO
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | 3-fluorobenzenesulfonamide, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-04-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
4V7H
 
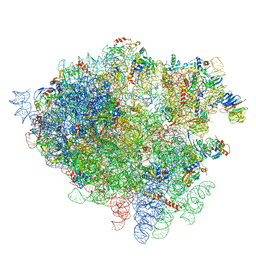 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
2X79
 
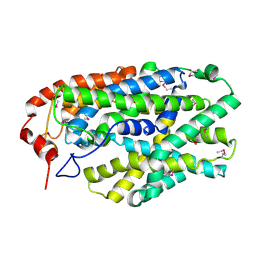 | | Inward facing conformation of Mhp1 | | Descriptor: | Hydantoin permease | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
3MJ5
 
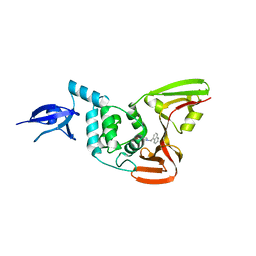 | | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation | | Descriptor: | N-(1,3-benzodioxol-5-ylmethyl)-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Replicase polyprotein 1a, ZINC ION | | Authors: | Mesecar, A.D, Ratia, K.M, Pegan, S.D. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation
J.Med.Chem., 53, 2010
|
|
2VH6
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with biaryl P4 motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-{(3S)-1-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]-2-oxopyrrolidin-3-yl}ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Chung, C.W, Convery, M.A, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A.N, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E. | | Deposit date: | 2007-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Biaryl P4 Motifs.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VS8
 
 | | The crystal structure of I-DmoI in complex with DNA and Mn | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP* GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*A)-3', ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WYJ
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
8FJH
 
 | | Crystal structure of RalA in a covalent complex with SOF-531 | | Descriptor: | 8-[bis(oxidanyl)-$l^{3}-sulfanyl]-~{N}-(3-fluoranyl-5-methoxy-phenyl)-2,3-dihydro-1,4-benzodioxine-5-sulfonamide, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Landgraf, A.D, Yeh, I.-J, Bum-Erdene, K, Gonzalez-Gutierrez, G, Meroueh, S. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring Covalent Bond Formation at Tyr-82 for Inhibition of Ral GTPase Activation.
Chemmedchem, 18, 2023
|
|
8FJI
 
 | | Crystal structure of RalA in a covalent complex with SOF-367 | | Descriptor: | 8-[fluoro(dihydroxy)-lambda~4~-sulfanyl]-N-(2-methoxypyridin-3-yl)-2,3-dihydro-1,4-benzodioxine-5-sulfonamide, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Landgraf, A.D, Yeh, I.-J, Bum-Erdene, K, Gonzalez-Gutierrez, G, Meroueh, S. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Exploring Covalent Bond Formation at Tyr-82 for Inhibition of Ral GTPase Activation.
Chemmedchem, 18, 2023
|
|
6UUG
 
 | | Structure of methanesulfinate monooxygenase MsuC from Pseudomonas fluorescens at 1.69 angstrom resolution | | Descriptor: | Putative dehydrogenase | | Authors: | Soule, J, Gnann, A.D, Gonzalez, R, Parker, M.J, McKenna, K.C, Nguyen, S.V, Phan, N.T, Wicht, D.K, Dowling, D.P. | | Deposit date: | 2019-10-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Structure and function of the two-component flavin-dependent methanesulfinate monooxygenase within bacterial sulfur assimilation.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
1DJS
 
 | |
8FFQ
 
 | | Wildtype rabbit TRPV5 into nanodiscs in the presence of PI(4,5)P2 and ruthenium red | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ERGOSTEROL, Transient receptor potential cation channel subfamily V member 5, ... | | Authors: | De Jesus-Perez, J.J, Fluck, E.C, Pumroy, R.A, Protopopova, A.D, Rocereta, J.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular details of ruthenium red pore block in TRPV channels.
Embo Rep., 25, 2024
|
|
8FFM
 
 | | Wildtype rat TRPV2 in nanodiscs bound to RR and 2-APB | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Pumroy, R.A, Protopopova, A.D, Rocereta, J.A, De Jesus-Perez, J.J, Fluck, E.C, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular details of ruthenium red pore block in TRPV channels.
Embo Rep., 25, 2024
|
|
