7YIR
 
 | |
7YIS
 
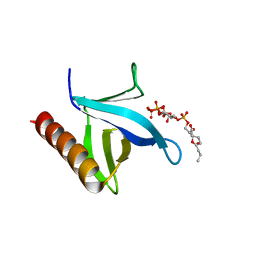 | | Crystal structure of N-terminal PH domain of ARAP3 protein in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Zhang, Y.J, Liu, Y.R, Wu, B. | | Deposit date: | 2022-07-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3.
Int J Mol Sci, 24, 2023
|
|
6K8K
 
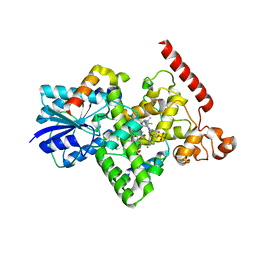 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5KAE
 
 | |
6JYT
 
 | |
8S5I
 
 | |
8S5L
 
 | |
8S5J
 
 | |
8S5K
 
 | |
8S5H
 
 | |
8S5M
 
 | |
6K8I
 
 | | Crystal structure of Arabidopsis thaliana CRY2 | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, L, Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KTO
 
 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | Descriptor: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6KA6
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 1 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-21 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
6KCN
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 4 | | Descriptor: | 3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)isochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-28 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
6KCT
 
 | |
6KBF
 
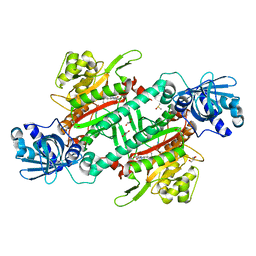 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 3 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydro-2~{H}-isoquinolin-1-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
7Y3I
 
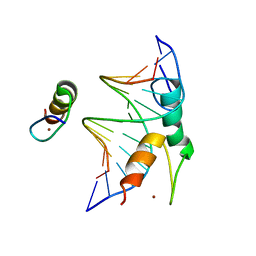 | | Structure of DNA bound SALL4 | | Descriptor: | DNA (12-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3L
 
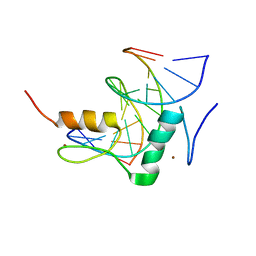 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | Descriptor: | DNA (12-mer), Sal-like protein 3, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
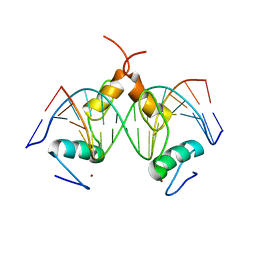 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3K
 
 | | Structure of SALL4 ZFC4 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
