1J9G
 
 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris S64C Mutant, monomer oxidised, at 2.4 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-25 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6I7E
 
 | | Plasmodium falciparum Myosin A, Pre-powerstroke | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-A, ... | | Authors: | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
5HMR
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor 3FMTDZ | | Descriptor: | 1-(1,2,3-thiadiazol-5-yl)-3-[3-(trifluoromethoxy)phenyl]urea, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Koncitikova, R, Briozzo, P. | | Deposit date: | 2016-01-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel thidiazuron-derived inhibitors of cytokinin oxidase/dehydrogenase.
Plant Mol.Biol., 92, 2016
|
|
1K2E
 
 | | crystal structure of a nudix protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6HHT
 
 | | Echovirus 18 Open particle without two pentamers | | Descriptor: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
5WDY
 
 | |
1J9E
 
 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris S35C Mutant at 1.44 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-25 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5MI8
 
 | | Structure of the phosphomimetic mutant of EF-Tu T383E | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
6HUG
 
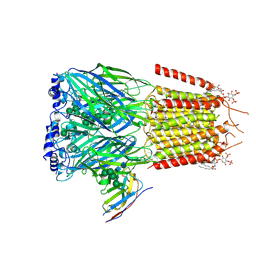 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with picrotoxin and megabody Mb38. | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
6HUO
 
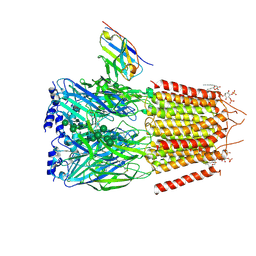 | | CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with alprazolam (Xanax), GABA and megabody Mb38. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloro-1-methyl-6-phenyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepine, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
1JN9
 
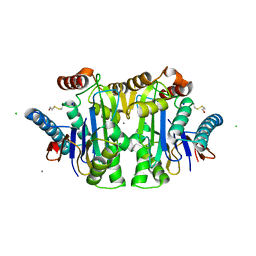 | |
1JOC
 
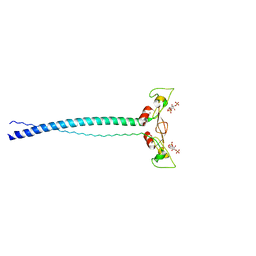 | | EEA1 homodimer of C-terminal FYVE domain bound to inositol 1,3-diphosphate | | Descriptor: | Early Endosomal Autoantigen 1, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ZINC ION | | Authors: | Dumas, J.J, Merithew, E, Rajamani, D, Hayes, S, Lawe, D, Corvera, S, Lambright, D.G. | | Deposit date: | 2001-07-27 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multivalent endosome targeting by homodimeric EEA1.
Mol.Cell, 8, 2001
|
|
1K26
 
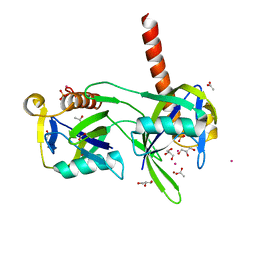 | | Structure of a Nudix Protein from Pyrobaculum aerophilum Solved by the Single Wavelength Anomolous Scattering Method | | Descriptor: | ACETIC ACID, GLYCEROL, IRIDIUM (III) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JUI
 
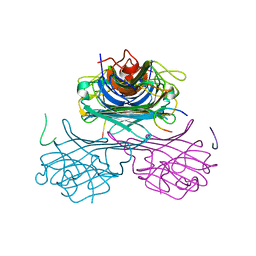 | | CONCANAVALIN A-CARBOHYDRATE MIMICKING 10-MER PEPTIDE COMPLEX | | Descriptor: | 10-mer Peptide, CALCIUM ION, Concanavalin-Br, ... | | Authors: | Jain, D, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2001-08-24 | | Release date: | 2002-08-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Plasticity in protein-peptide recognition: crystal structures of two different peptides bound to concanavalin A.
Biophys.J., 80, 2001
|
|
6QUL
 
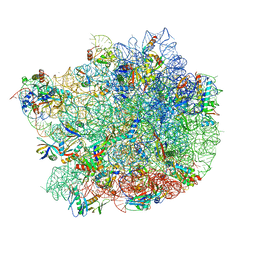 | | Structure of a bacterial 50S ribosomal subunit in complex with the novel quinoxolidinone antibiotic cadazolid | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Scaiola, A, Leibundgut, M, Boehringer, D, Ritz, D. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of translation inhibition by cadazolid, a novel quinoxolidinone antibiotic.
Sci Rep, 9, 2019
|
|
9FUW
 
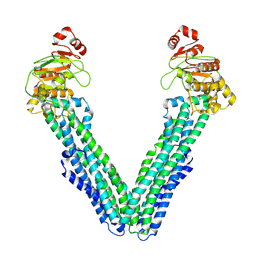 | | MsbA in Triton inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV8
 
 | | MsbA in Amphipol 18 inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
5LFF
 
 | | NMR structure of peptide 2 targeting CXCR4 | | Descriptor: | ARG-ALA-CYS-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
9FUZ
 
 | | MsbA in UDM inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FVA
 
 | | MsbA in MSP2N2 Nanodisc inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUU
 
 | | MsbA in GDN inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV7
 
 | | MsbA in Amphipol A8-35 inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
7AAP
 
 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1FIA
 
 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
7AA5
 
 | | Human TRPV4 structure in presence of 4a-PDD | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | Authors: | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
