4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HEC
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
5C56
 
 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
2HOB
 
 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | Authors: | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
5YQZ
 
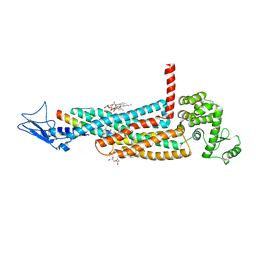 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
1LBW
 
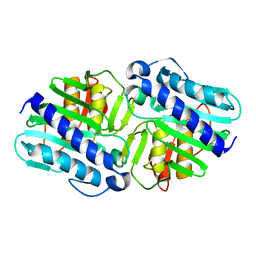 | | Crystal Structure of apo-form (P32) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBX
 
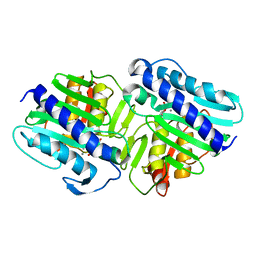 | | Crystal Structure of a ternary complex of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with Calcium ions and D-myo-Inositol-1-Phosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1-PHOSPHATE, fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBZ
 
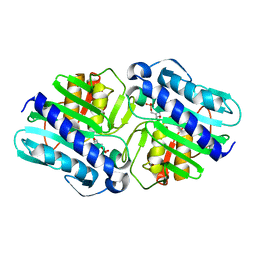 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Calcium ions and Fructose-1,6 bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
6LND
 
 | | Crystal structure of transposition protein TniQ | | Descriptor: | ZINC ION, transposition protein TniQ | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
5U78
 
 | | Crystal structure of ORP8 PH domain in P1211 space group | | Descriptor: | Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
5U77
 
 | | Crystal structure of ORP8 PH domain | | Descriptor: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
8HNQ
 
 | | The structure of a alcohol dehydrogenase AKR13B2 with NADP | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chen, M, Yang, H, Lu, F. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of AKR13B2
To Be Published
|
|
8HW0
 
 | | the structure of AKR6D1 | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent aldo/keto reductase AKR6D1 | | Authors: | Chen, M, Yang, H. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | structure of AKR6D1
To Be Published
|
|
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | Descriptor: | DNA-binding protein Alba | | Authors: | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | Deposit date: | 2002-12-19 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
8H01
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8H00
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8GZZ
 
 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | Descriptor: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
2Q62
 
 | | Crystal Structure of ArsH from Sinorhizobium meliloti | | Descriptor: | SULFATE ION, arsH | | Authors: | Ye, J, Yang, H, Bhattacharjee, H, Rosen, B.P. | | Deposit date: | 2007-06-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti
FEBS Lett., 581, 2007
|
|
2PH7
 
 | | Crystal structure of AF2093 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_2093 | | Authors: | Chang, J.C, Yang, H, Hwang, J, Zhu, J, Chen, L, Fu, Z.-Q, Xu, H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AF2093 from Archaeoglobus fulgidus.
To be Published
|
|
4PXZ
 
 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
