8E58
 
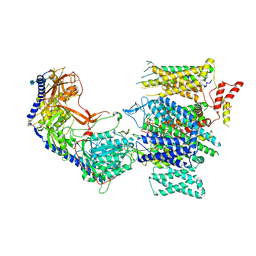 | | Rabbit L-type voltage-gated calcium channel Cav1.1 in the presence of Amiodarone and 1 mM MNI-1 at 3.0 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
6K7P
 
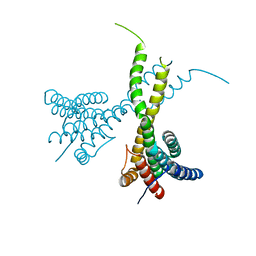 | | Crystal structure of human AFF4-THD domain | | Descriptor: | AF4/FMR2 family member 4 | | Authors: | Tang, D, Xue, Y, Li, S, Cheng, W, Duan, J, Wang, J, Qi, S. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into the effect of AFF4 dimerization on activation of HIV-1 proviral transcription.
Cell Discov, 6, 2020
|
|
8ZF1
 
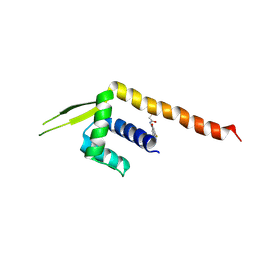 | | Crystal structure of a Chemo Triplet Photoenzyme (CTPe) | | Descriptor: | N-(9-oxidanylidenethioxanthen-2-yl)ethanamide, Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Guo, J, Wu, Y.Z, Zhong, F.R. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemogenetic Evolution of Diversified Photoenzymes for Enantioselective [2 + 2] Cycloadditions in Whole Cells.
J.Am.Chem.Soc., 2024
|
|
6NZ7
 
 | |
5IL0
 
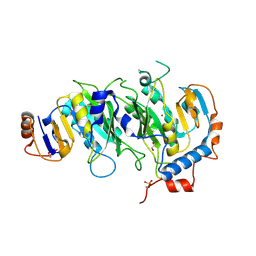 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL1
 
 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5JO5
 
 | |
5JNY
 
 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
8F60
 
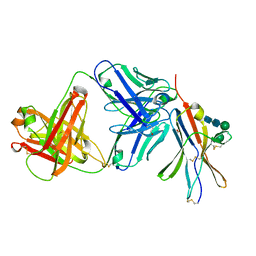 | | anti-BTLA monoclonal antibody r23C8 in complex with BTLA | | Descriptor: | ACETATE ION, B- and T-lymphocyte attenuator, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6O
 
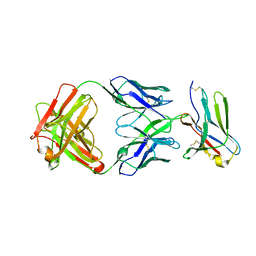 | | anti-BTLA monoclonal antibody h22B3 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h22B3 Fab heavy chain, h22B3 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6L
 
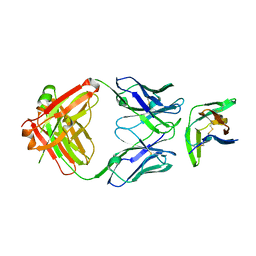 | | anti-BTLA monoclonal antibody h25F7 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h25F7 Fab heavy chain, h25F7 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
5JR1
 
 | |
7CMA
 
 | | Structure of A151R from African swine fever virus Georgia | | Descriptor: | A151R, ZINC ION | | Authors: | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | Deposit date: | 2020-07-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
5V0Y
 
 | | Solution structure of arenicin-3. | | Descriptor: | arenicin-3 | | Authors: | Edwards, I.A, Mobli, M. | | Deposit date: | 2017-02-28 | | Release date: | 2018-08-08 | | Last modified: | 2020-06-10 | | Method: | SOLUTION NMR | | Cite: | An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria
Nat Commun, 2020
|
|
7LZ4
 
 | | Crystal structure of A211D mutant of Protein Kinase A RIa subunit, a Carney Complex mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed | | Authors: | Del Rio, J, Wu, J, Taylor, S.S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.155 Å) | | Cite: | Noncanonical protein kinase A activation by oligomerization of regulatory subunits as revealed by inherited Carney complex mutations.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8HL9
 
 | | Crystal structure of Galectin-8 C-CRD with part of linker | | Descriptor: | Galectin-8 | | Authors: | Si, Y.L. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Linker remodels human Galectin-8 structure and regulates its hemagglutination and pro-apoptotic activity.
Int.J.Biol.Macromol., 245, 2023
|
|
3N2R
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R/3S,4S)-4-(3-Phenoxyphenoxy)pyrrolidin-3-yl)methyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(3-phenoxyphenoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2010-05-18 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of lipophilic-tailed monocationic inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 18, 2010
|
|
7V1N
 
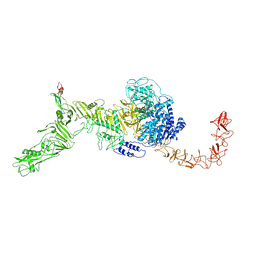 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
5DIH
 
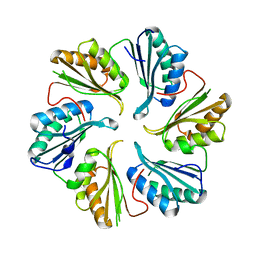 | |
5DII
 
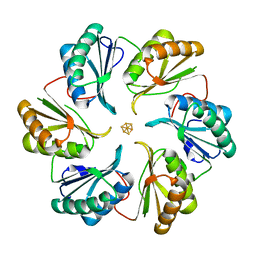 | | Structure of an engineered bacterial microcompartment shell protein binding a [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, Microcompartments protein | | Authors: | Sutter, M, Aussignargues, C, Turmo, A, Kerfeld, C.A. | | Deposit date: | 2015-09-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster.
J.Am.Chem.Soc., 138, 2016
|
|
7Y99
 
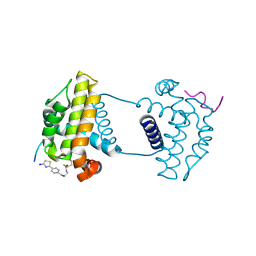 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y90
 
 | | Crystal Structure Analysis of cp1 bound BCL2 | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W. | | Deposit date: | 2022-06-24 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
