8TEP
 
 | | Human cytomegalovirus portal vertex, virion configuration 1 (VC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Inner tegument protein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEU
 
 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 2 - inverted (NC2-inv) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
6UX2
 
 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PQ5
 
 | |
6PQA
 
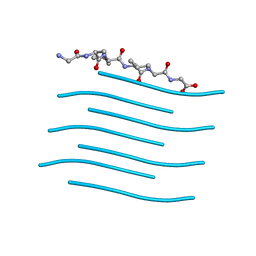 | |
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UZB
 
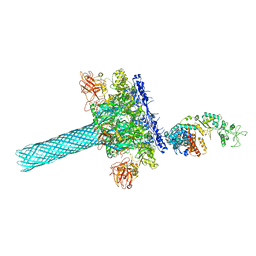 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6UZE
 
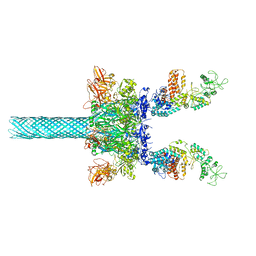 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6UZD
 
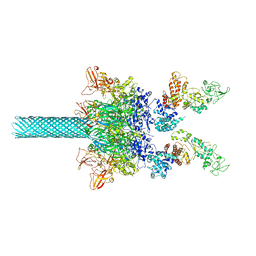 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
5LBV
 
 | | Structural basis of zika and dengue virus potent antibody cross-neutralization | | Descriptor: | SODIUM ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, envelope protein E | | Authors: | Barba-Spaeth, G. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5LBS
 
 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5LCV
 
 | |
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8EEQ
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-09-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8OKH
 
 | |
6V0V
 
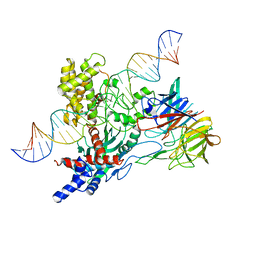 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7UAK
 
 | |
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
5IKL
 
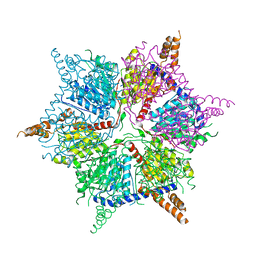 | |
5UBO
 
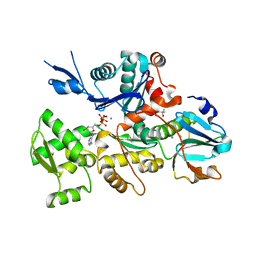 | | Mical-oxidized Actin complex with Gelsolin Segment 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Grintsevich, E.E, Sawaya, M.R, Reisler, E. | | Deposit date: | 2016-12-21 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
5W50
 
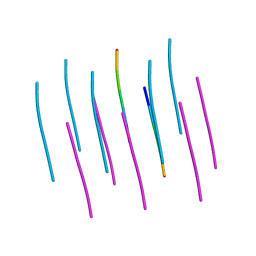 | | Crystal structure of the segment, LIIKGI, from the RRM2 of TDP-43, residues 248-253 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-06-13 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5W0K
 
 | |
5W52
 
 | | MicroED structure of the segment, DLIIKGISVHI, from the RRM2 of TDP-43, residues 247-257 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2017-06-13 | | Release date: | 2018-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3U9T
 
 | |
