2RB6
 
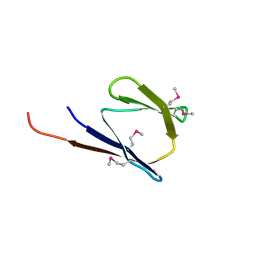 | | X-Ray structure of the protein Q8EI81. Northeast Structural Genomics Consortium target SoR78A | | Descriptor: | Uncharacterized protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Vorobiev, S.M, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the protein Q8EI81.
To be Published
|
|
2RAL
 
 | | Crystal Structure Analysis of double cysteine mutant of S.epidermidis adhesin SdrG: Evidence for the Dock,Lock and Latch ligand binding mechanism | | Descriptor: | Serine-aspartate repeat-containing protein G | | Authors: | Ponnuraj, K, Sthanam, N, Bowden, M.G, Hook, M. | | Deposit date: | 2007-09-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the "dock, lock, and latch" ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG.
J.Biol.Chem., 283, 2008
|
|
4URY
 
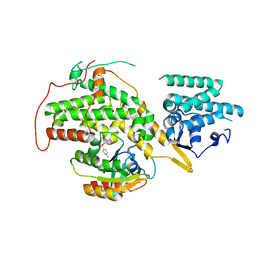 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | GTPASE HRAS, N-[(4-aminophenyl)sulfonyl]cyclopropanecarboxamide, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
2RD4
 
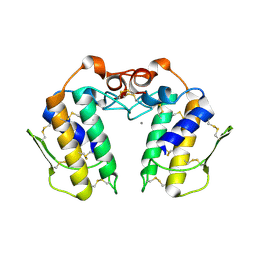 | | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 1, Phospholipase A2 isoform 2, ... | | Authors: | Mirza, Z, Kaur, A, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution
To be Published
|
|
4V0T
 
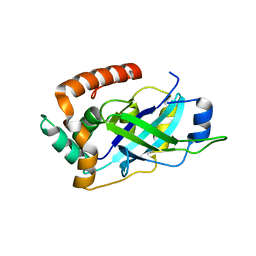 | | Monomeric pseudorabies virus protease pUL26N at 2.1 A resolution | | Descriptor: | UL26 | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
4V4T
 
 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4W6W
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF6 | | Descriptor: | F18 fimbrial adhesin AC, NbFedF6 | | Authors: | Moonens, K, De Kerpel, M, Annelies, C, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
2RK4
 
 | | Structure of M26I DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
2R6T
 
 | | Structure of a R132K variant PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobalamin adenosyltransferase PduO-like protein, MAGNESIUM ION | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional analyses of the human-type corrinoid adenosyltransferase (PduO) from Lactobacillus reuteri.
Biochemistry, 46, 2007
|
|
2RM4
 
 | |
2RO3
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator Abh | | Descriptor: | Putative transition state regulator abh | | Authors: | Sullivan, D.M, Bobay, B.G, Douglas, K.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
4V69
 
 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4W6X
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF7 | | Descriptor: | F18 fimbrial adhesin AC, Nanobody NbFedF7 | | Authors: | Moonens, K, De Kerpel, M, Coddens, A, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
2RF8
 
 | |
2R98
 
 | | Crystal Structure of N-acetylglutamate synthase (selenoMet substituted) from Neisseria gonorrhoeae | | Descriptor: | ACETYL COENZYME *A, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
2R9G
 
 | | Crystal structure of the C-terminal fragment of AAA ATPase from Enterococcus faecium | | Descriptor: | AAA ATPase, central region, ACETATE ION, ... | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Shi, W, Toro, R, Meyer, A.J, Rutter, M, Wu, B, Groshong, C, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-12 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of AAA ATPase from Enterococcus faecium.
To be Published
|
|
4WBA
 
 | | Q/E mutant SA11 NSP4_CCD | | Descriptor: | GLYCEROL, Non-structural glycoprotein NSP4, PHOSPHATE ION | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
2RLC
 
 | |
2RLT
 
 | | phosphorylated CPI-17 (22-120) | | Descriptor: | Protein phosphatase 1 regulatory subunit 14A | | Authors: | Eto, M. | | Deposit date: | 2007-08-11 | | Release date: | 2008-07-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor.
Structure, 15, 2007
|
|
2RM5
 
 | | Glutathione peroxidase-type tryparedoxin peroxidase, oxidized form | | Descriptor: | Glutathione peroxidase-like protein | | Authors: | Melchers, J, Feher, K, Diechtierow, M, Krauth-Siegel, L, Tews, I, Muhle-Goll, C. | | Deposit date: | 2007-10-03 | | Release date: | 2008-07-29 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural basis for a distinct catalytic mechanism in Trypanosoma brucei tryparedoxin peroxidase
J.Biol.Chem., 283, 2008
|
|
2REX
 
 | | Crystal structure of the effector domain of PLXNB1 bound with Rnd1 GTPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tong, Y, Tempel, W, Shen, L, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-27 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the effector domain of PLXNB1 bound with Rnd1 GTPase.
To be Published
|
|
4WB5
 
 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
2RRD
 
 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
4WBH
 
 | |
