3S3G
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Tolmetin | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tolmetin | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
2PNU
 
 | | Crystal structure of human androgen receptor ligand-binding domain in complex with EM-5744 | | Descriptor: | (5S,8R,9S,10S,13R,14S,17S)-13-{2-[(3,5-DIFLUOROBENZYL)OXY]ETHYL}-17-HYDROXY-10-METHYLHEXADECAHYDRO-3H-CYCLOPENTA[A]PHENANTHREN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Cantin, L, Faucher, F, Couture, J.F, Pereira de Jesus-Tran, K, Legrand, P, Ciobanu, C.L, Singh, S.M, Labrie, F, Breton, R. | | Deposit date: | 2007-04-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
J.Biol.Chem., 282, 2007
|
|
7BTN
 
 | |
6JBX
 
 | | Crystal structure of Streptococcus pneumoniae FabT in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*GP*TP*TP*TP*GP*AP*CP*TP*GP*TP*CP*AP*AP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*AP*AP*TP*TP*TP*GP*AP*CP*AP*GP*TP*CP*AP*AP*AP*CP*TP*AP*TP*T)-3'), Fatty acid biosynthesis transcriptional regulator, ... | | Authors: | Zuo, G, Chen, Z.P, Li, Q, Zhou, C.Z. | | Deposit date: | 2019-01-27 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into repression of the Pneumococcal fatty acid synthesis pathway by repressor FabT and co-repressor acyl-ACP.
Febs Lett., 593, 2019
|
|
1SHC
 
 | | SHC PTB DOMAIN COMPLEXED WITH A TRKA RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SHC, TRKA RECEPTOR PHOSPHOPEPTIDE | | Authors: | Zhou, M.-M, Ravichandran, K.S, Olejniczak, E.T, Petros, A.M, Meadows, R.P, Sattler, M, Harlan, J.E, Wade, W.S, Burakoff, S.J, Fesik, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Nature, 378, 1995
|
|
7CMO
 
 | | Crystal structure of human inorganic pyrophosphatase | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Hu, F, Huang, Z, Li, L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biochemical characterization of inorganic pyrophosphatase from Homo sapiens.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
3GBI
 
 | | The Rational Design and Structural Analysis of a Self-Assembled Three-Dimensional DNA Crystal | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Birktoft, J.J, Zheng, J, Seeman, N.C. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.018 Å) | | Cite: | From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal.
Nature, 461, 2009
|
|
7CQZ
 
 | | crystal structure of mouse FAM46C (TENT5C) | | Descriptor: | CHLORIDE ION, GLYCEROL, Terminal nucleotidyltransferase 5C | | Authors: | Hu, J.L, Zhang, H, Gao, S. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34969759 Å) | | Cite: | Structural and functional characterization of multiple myeloma associated cytoplasmic poly(A) polymerase FAM46C.
Cancer Commun (Lond), 41, 2021
|
|
2FVD
 
 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
2H0Q
 
 | |
3JAF
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.801 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
4P7H
 
 | |
6KZK
 
 | |
3JAD
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, strychnine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, STRYCHNINE | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3HR5
 
 | |
1TZH
 
 | |
1TZI
 
 | |
3JAE
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
6KCW
 
 | | Structure of alginate lyase Aly36B | | Descriptor: | Alginate lyase, CALCIUM ION, PHOSPHATE ION | | Authors: | Dong, F, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
7EEA
 
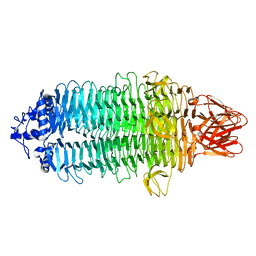 | |
7EEQ
 
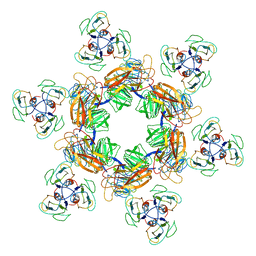 | | Cyanophage Pam1 tail machine | | Descriptor: | Needle head proteins, Tailspike head-binding domain | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEP
 
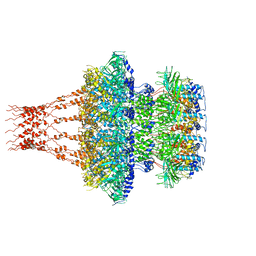 | | Cyanophage Pam1 portal-adaptor complex | | Descriptor: | Pam1 adaptor proteins, Pam1 portal proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEL
 
 | | Cyanophage Pam1 capsid asymmetric unit | | Descriptor: | Cement (decoration) proteins, Major capsid proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
6KBW
 
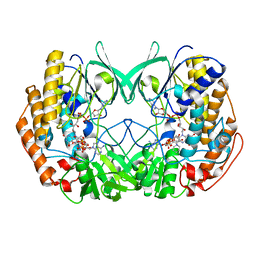 | | Crystal structure of Tmm from Myroides profundi D25 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethylamine monooxygenase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Oxidation of trimethylamine to trimethylamine N-oxide facilitates high hydrostatic pressure tolerance in a generalist bacterial lineage.
Sci Adv, 7, 2021
|
|
