3JRB
 
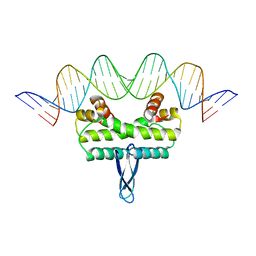 | |
3JRH
 
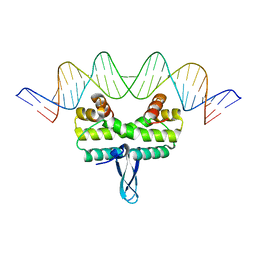 | |
4I7A
 
 | | GrpN pentameric microcompartment shell protein from Rhodospirillum rubrum | | Descriptor: | CHLORIDE ION, Ethanolamine utilization protein EutN/carboxysome structural protein Ccml | | Authors: | Wheatley, N.M, Gidaniyan, S.D, Cascio, D, Yeates, T.O. | | Deposit date: | 2012-11-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins.
Protein Sci., 22, 2013
|
|
3JRC
 
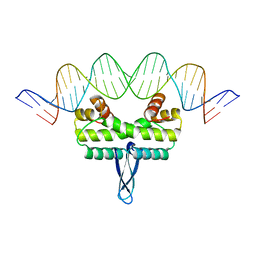 | |
3JRI
 
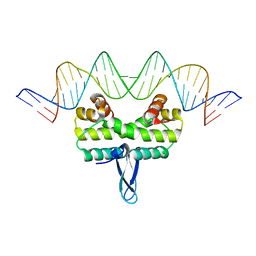 | |
3HWL
 
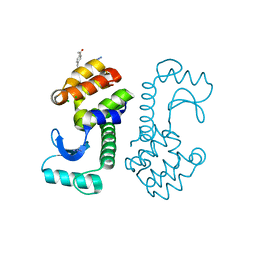 | | Crystal Structure of T4 lysozyme with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 131 | | Descriptor: | AZIDE ION, CHLORIDE ION, Lysozyme | | Authors: | Fleissner, M.R, Cascio, D, Schultz, P.G, Hubbell, W.L. | | Deposit date: | 2009-06-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed spin labeling of a genetically encoded unnatural amino acid.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3JRG
 
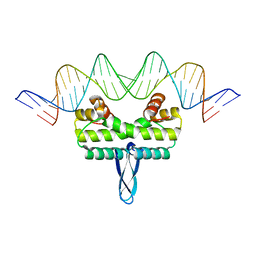 | |
3JRE
 
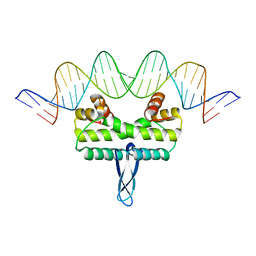 | |
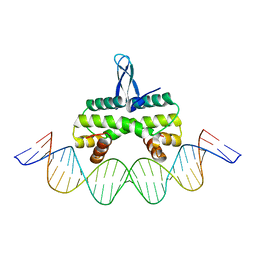
4IHV
 
 | |
3LSU
 
 | |
3NGK
 
 | |
3DH4
 
 | | Crystal Structure of Sodium/Sugar symporter with bound Galactose from vibrio parahaemolyticus | | Descriptor: | ERBIUM (III) ION, SODIUM ION, Sodium/glucose cotransporter, ... | | Authors: | Abramson, J, Faham, S, Cascio, D. | | Deposit date: | 2008-06-16 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport.
Science, 321, 2008
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4GUN
 
 | |
4IBL
 
 | | Rubidium Sites in Blood Coagulation Factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schmidt, A, Cascio, D, Padmanabhan, K, Bajaj, S.P. | | Deposit date: | 2012-12-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human factor VIIa-soluble tissue factor with calcium, magnesium and rubidium
Acta Crystallogr.,Sect.D, D77, 2021
|
|
3N79
 
 | | PduT C38S Mutant from Salmonella enterica Typhimurium | | Descriptor: | CHLORIDE ION, PduT, SODIUM ION, ... | | Authors: | Crowley, C.S, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into the Mechanisms of Transport across the Salmonella enterica Pdu Microcompartment Shell.
J.Biol.Chem., 285, 2010
|
|
3NG0
 
 | |
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RPM
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM with bound hexanoyl | | Descriptor: | HEXANOIC ACID, HEXANOYL-COENZYME A, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RUB
 
 | | A CRYSTAL FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM NICOTIANA TABACUM IN THE ACTIVATED STATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Eisenberg, D. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal form of ribulose-1,5-bisphosphate carboxylase/oxygenase from Nicotiana tabacum in the activated state.
J.Mol.Biol., 197, 1987
|
|
3SGN
 
 | |
3Q28
 
 | | Cyrstal structure of human alpha-synuclein (58-79) fused to maltose binding protein (MBP) | | Descriptor: | Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q29
 
 | | Cyrstal structure of human alpha-synuclein (1-19) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3SGP
 
 | | Amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4016 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
