1QFD
 
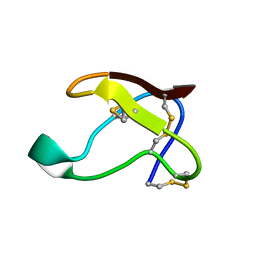 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | Descriptor: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | Authors: | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | Deposit date: | 1999-04-08 | | Release date: | 1999-07-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
1QK7
 
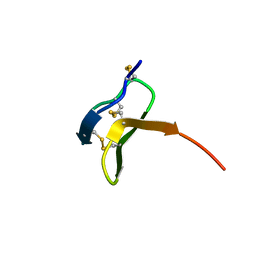 | |
3DV2
 
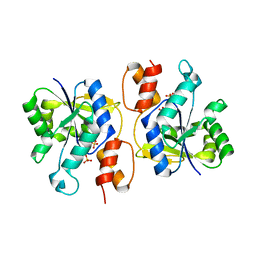 | | Crystal Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis | | Descriptor: | Nicotinate (Nicotinamide) nucleotide adenylyltransferase, SULFATE ION | | Authors: | Lu, S, Smith, C.D, Yang, Z, Pruett, P.S, Nagy, L, McCombs, D.P, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
7MJ5
 
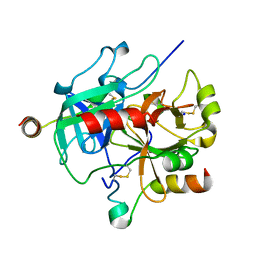 | | complex of human thrombin with XC-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Putative secreted salivary protein, SODIUM ION, ... | | Authors: | Lu, S, Tirloni, L, Andersen, J.F. | | Deposit date: | 2021-04-19 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of a substrate-like cleavage-resistant thrombin inhibitor from the saliva of the flea Xenopsylla cheopis.
J.Biol.Chem., 297, 2021
|
|
8DOD
 
 | | Beta-lactamase CTX-M-14 S130A | | Descriptor: | Beta-lactamase, POTASSIUM ION | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-12 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DOE
 
 | | Crystal Structure of CTX-M-14 N106A | | Descriptor: | Beta-lactamase | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-12 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DP4
 
 | | Beta-lactamase CTX-M-14 T235A | | Descriptor: | Beta-lactamase | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DON
 
 | | Beta-lactamase CTX-M-14 T215A | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DPQ
 
 | | Beta-lactamase CTX-M-14 N170A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Lu, S, Neetu, N, Palzkill, T. | | Deposit date: | 2022-07-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8ELA
 
 | | CTX-M-14 beta-lactamase mutant - N132A w MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lu, S, Palzkill, T, Hu, L, Prasad, B.V.V. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8ELB
 
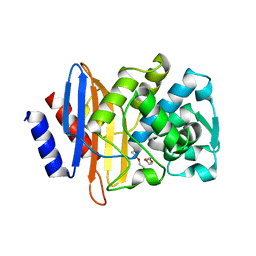 | | CTX-M-14 beta-lactamase mutant- N132A | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER | | Authors: | Lu, S, Neetu, N, Palzkill, T. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
7S5S
 
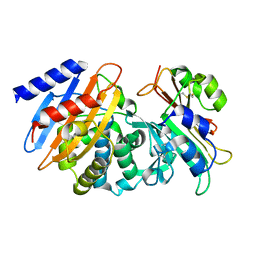 | | CTX-M-15 WT in complex with BLIP WT | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein | | Authors: | Lu, S, Palzkill, T, Hu, L.Y, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2021-09-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An active site loop toggles between conformations to control antibiotic hydrolysis and inhibition potency for CTX-M beta-lactamase drug-resistance enzymes.
Nat Commun, 13, 2022
|
|
8GDL
 
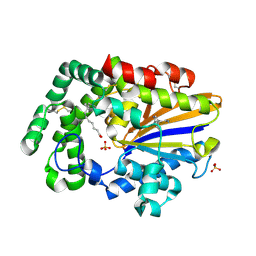 | | Acid phosphatase pseudoenzyme from flea | | Descriptor: | PALMITOLEIC ACID, SEROTONIN, SULFATE ION, ... | | Authors: | Lu, S, Andersen, J.F, Ribeiro, J.M. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Acid phosphatase-like proteins, a biogenic amine and leukotriene-binding salivary protein family from the flea Xenopsylla cheopis.
Commun Biol, 6, 2023
|
|
7LTN
 
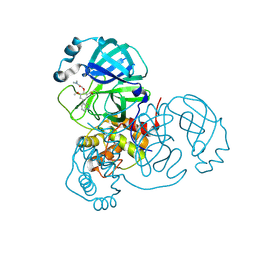 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | Descriptor: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
7K2X
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase | | Descriptor: | Beta-lactamase, GLYCEROL | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7K2W
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7K2Y
 
 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7US4
 
 | | Sars-Cov2 Main Protease in complex with CDD-1819 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7UR9
 
 | | SARS-Cov2 Main protease in complex with inhibitor CDD-1845 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[4-(methylamino)-4-oxobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7URB
 
 | | Sars-Cov2 Main Protease in complex with CDD-1733 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-{(1S)-1-[4-(trifluoromethyl)phenyl]butyl}-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
1QWK
 
 | | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa protein (aldose reductase family member) | | Descriptor: | aldo-keto reductase family 1 member C1 | | Authors: | Chen, L, Zhou, X.E, Meehan, E.J, Symersky, J, Lu, S, Li, S, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-09-02 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa
protein (aldose reductase family member)
To be published
|
|
1XHL
 
 | | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase family member (5L265), ... | | Authors: | Schormann, N, Karpova, E, Zhou, J, Zhang, Y, Symersky, J, Bunzel, R, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKInstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate
To be Published
|
|
1XKQ
 
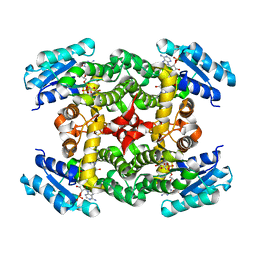 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
1YIS
 
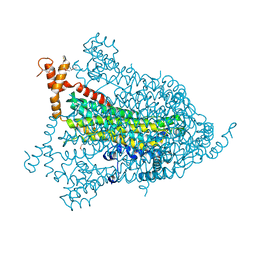 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
