2M0M
 
 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
2MT3
 
 | |
4LX3
 
 | | Conserved Residues that Modulate Protein trans-Splicing of Npu DnaE Split Intein | | Descriptor: | DNA polymerase III, alpha subunit, Nucleic acid binding, ... | | Authors: | Wu, Q, Gao, Z, Wei, Y, Ma, G, Zheng, Y, Dong, Y, Liu, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved residues that modulate protein trans-splicing of Npu DnaE split intein.
Biochem.J., 461, 2014
|
|
4MMJ
 
 | | crystal structure of YafQ from E.coli strain BL21(DE3) | | Descriptor: | Addiction module toxin, RelE/StbE family, SULFATE ION | | Authors: | Liang, Y, Gao, Z, Liu, Q, Dong, Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4XO1
 
 | |
6LB3
 
 | | Crystal structure of PA4674 in complex with its operator DNA (18bp) from Pseudomonas aeruginosa | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*TP*TP*AP*AP*CP*GP*TP*TP*AP*AP*GP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*CP*TP*TP*AP*AP*CP*GP*TP*TP*AP*AP*GP*GP*GP*T)-3'), HTH cro/C1-type domain-containing protein, ... | | Authors: | Liu, Y, Zhang, H, Gao, Z, Dong, Y. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structure of PA4674 in complex with its operator DNA (18bp) from Pseudomonas aeruginosa
To Be Published
|
|
4ZW0
 
 | |
2MA1
 
 | | Solution structure of HRDC1 domain of RecQ helicase from Deinococcus radiodurans | | Descriptor: | DNA helicase RecQ | | Authors: | Liu, S, Zhang, W, Gao, Z, Ming, Q, Hou, H, Lan, W, Wu, H, Cao, C, Dong, Y. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal-most HRDC1 domain of RecQ helicase from Deinococcus radiodurans
To be Published
|
|
1BJH
 
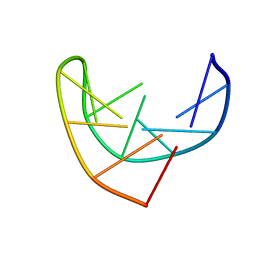 | | HAIRPIN LOOPS CONSISTING OF SINGLE ADENINE RESIDUES CLOSED BY SHEARED A(DOT)A AND G(DOT)G PAIRS FORMED BY THE DNA TRIPLETS AAA AND GAG: SOLUTION STRUCTURE OF THE D(GTACAAAGTAC) HAIRPIN, NMR, 16 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*AP*AP*AP*GP*TP*AP*C)-3') | | Authors: | Chou, S.-H, Zhu, L, Gao, Z, Cheng, J.-W, Reid, B.R. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin.
J.Mol.Biol., 264, 1996
|
|
6M10
 
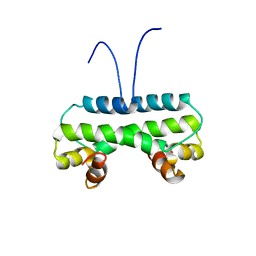 | | Crystal structure of PA4853 (Fis) from Pseudomonas aeruginosa | | Descriptor: | Putative Fis-like DNA-binding protein | | Authors: | Zhang, H, Gao, Z, Zhou, J, Dong, Y. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of the nucleoid-associated protein Fis (PA4853) from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4FVD
 
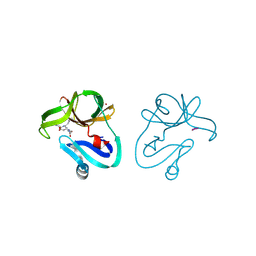 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4FVB
 
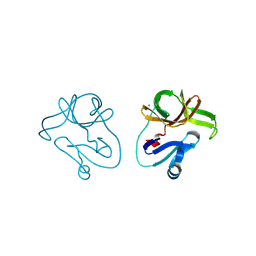 | | Crystal structure of EV71 2A proteinase C110A mutant | | Descriptor: | 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
6JPI
 
 | |
5YOX
 
 | | HD domain-containing protein YGK1(YGL101W) | | Descriptor: | HD domain-containing protein YGL101W, ZINC ION | | Authors: | Yang, J, Wang, F, Gao, Z, Zhou, K, Liu, Q. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | HD domain-containing protein YGK1(YGL101W)
To Be Published
|
|
6ITW
 
 | |
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | Descriptor: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
4W7T
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4MMG
 
 | | crystal structure of YafQ mutant H87Q from E.coli | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4ML0
 
 | | Crystal structure of E.coli DinJ-YafQ complex | | Descriptor: | Predicted antitoxin of YafQ-DinJ toxin-antitoxin system, Predicted toxin of the YafQ-DinJ toxin-antitoxin system, SULFATE ION | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4ML2
 
 | | Crystal structure of wild-type YafQ | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
5IXY
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 31: (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one | | Descriptor: | (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chen, Z, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
5IXS
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 9: (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one | | Descriptor: | (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
4XO2
 
 | |
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
