2EW7
 
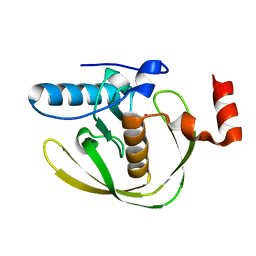 | | Crystal Structure of Helicobacter Pylori peptide deformylase | | Descriptor: | COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW6
 
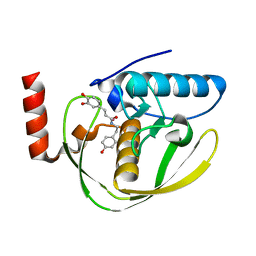 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | (2E)-3-(3,4-DIHYDROXYPHENYL)-N-[2-(4-HYDROXYPHENYL)ETHYL]ACRYLAMIDE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW5
 
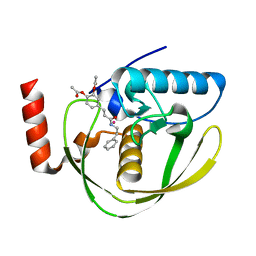 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | 4-{(1E)-3-OXO-3-[(2-PHENYLETHYL)AMINO]PROP-1-EN-1-YL}-1,2-PHENYLENE DIACETATE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2LKK
 
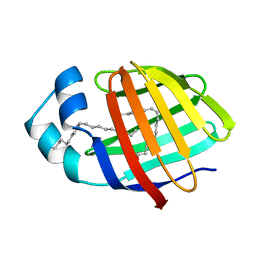 | | Human L-FABP in complex with oleate | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2011-10-13 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2L67
 
 | | Solution Structure of Human Apo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2L68
 
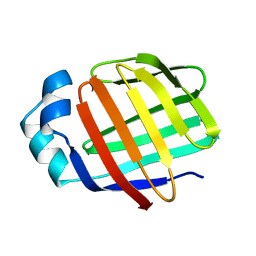 | | Solution Structure of Human Holo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
8HLG
 
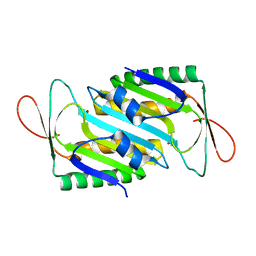 | | Crystal structure of MoaE | | Descriptor: | Molybdenum cofactor biosynthesis protein D/E, SULFATE ION | | Authors: | Cai, J, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans.
Int J Mol Sci, 24, 2023
|
|
3BJC
 
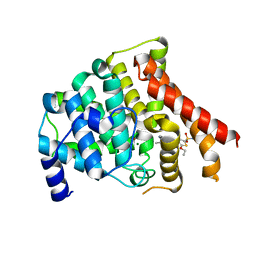 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | Descriptor: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | Deposit date: | 2007-12-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
3GLR
 
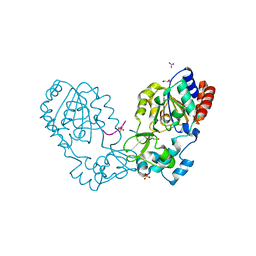 | | Crystal Structure of human SIRT3 with acetyl-lysine AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, BICARBONATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLT
 
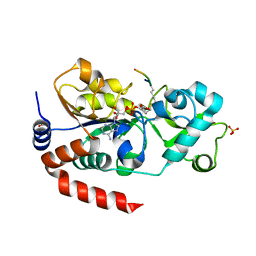 | | Crystal Structure of Human SIRT3 with ADPR bound to the AceCS2 peptide containing a thioacetyl lysine | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, CARBONATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
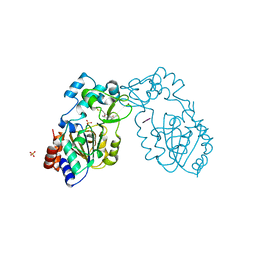
3ECN
 
 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
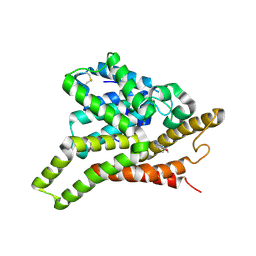
3GLU
 
 | | Crystal Structure of Human SIRT3 with AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, NAD-dependent deacetylase sirtuin-3, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3ECM
 
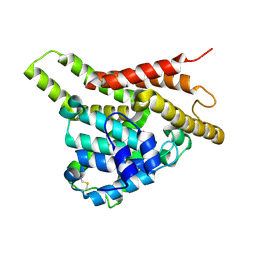 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3GLS
 
 | | Crystal Structure of Human SIRT3 | | Descriptor: | NAD-dependent deacetylase sirtuin-3, mitochondrial, SULFATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
2H40
 
 | | Crystal structure of the catalytic domain of unliganded PDE5 | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Liu, Y, Huai, Q, Cai, J, Zoraghi, R, Francis, S.H, Corbin, J.D, Robinson, H, Xin, Z, Lin, G, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
8CQY
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the PBM TACE C-terminal peptide | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
1HX3
 
 | | CRYSTAL STRUCTURE OF E.COLI ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | IMIDAZOLE, ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
8OEP
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the HPV18 E6 oncoprotein C-terminal peptide | | Descriptor: | Protein E6, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | Deposit date: | 2023-03-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
2BM2
 
 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | Descriptor: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | Authors: | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
1HZT
 
 | | CRYSTAL STRUCTURE OF METAL-FREE ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
1KOG
 
 | | Crystal structure of E. coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase, Threonyl-tRNA synthetase mRNA, ... | | Authors: | Torres-Larrios, A, Dock-Bregeon, A.C, Romby, P, Rees, B, Sankaranarayanan, R, Caillet, J, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat.Struct.Biol., 9, 2002
|
|
6Q56
 
 | | Crystal structure of the B. subtilis M1A22 tRNA methyltransferase TrmK | | Descriptor: | NICKEL (II) ION, tRNA (adenine(22)-N(1))-methyltransferase | | Authors: | Degut, C, Roovers, M, Barraud, P, Brachet, F, Feller, A, Larue, V, Al Refaii, A, Caillet, J, Droogmans, L, Tisne, C. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of B. subtilis m1A22 tRNA methyltransferase TrmK: insights into tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
1TKE
 
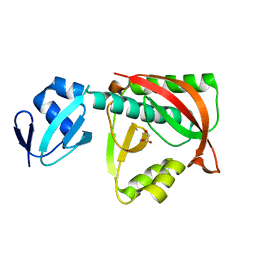 | | Crystal structure of the editing domain of threonyl-tRNA synthetase complexed with serine | | Descriptor: | SERINE, Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-08 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1TKG
 
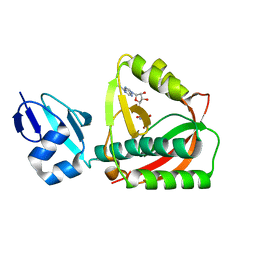 | | Crystal structure of the editing domain of threonyl-tRNA synthetase complexed with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-08 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1TKY
 
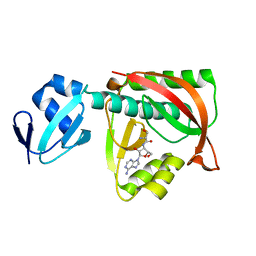 | | Crystal structure of the editing domain of threonyl-tRNA synthetase complexed with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
