2E8X
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and GPP | | Descriptor: | GERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Ko, T.P, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E93
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with BPH-629 | | Descriptor: | Geranylgeranyl pyrophosphate synthetase, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Guo, R.T, Cao, R, Ko, T.P, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E99
 
 | | E. coli undecaprenyl pyrophosphate synthase in complex with BPH-608 | | Descriptor: | (1-HYDROXY-1-PHOSPHONO-2-[1,1';3',1'']TERPHENYL-3-YL-ETHYL)-PHOSPHONIC ACID, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Liang, P.H, Oldfield, E, Wang, A.H.J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E8T
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium, FsPP and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION, ... | | Authors: | Guo, R.T, Ko, T.P, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8I9G
 
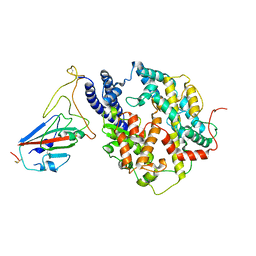 | | S-RBD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the enhanced infectivity and immune evasion of Omicron subvariants
To Be Published
|
|
8I9F
 
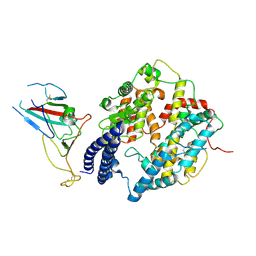 | | S-RBD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the enhanced infectivity and immune evasion of Omicron subvariants
To Be Published
|
|
8I9B
 
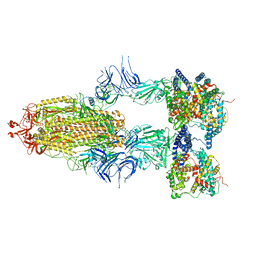 | | S-ECD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the enhanced infectivity and immune evasion of Omicron subvariants
To Be Published
|
|
5X5O
 
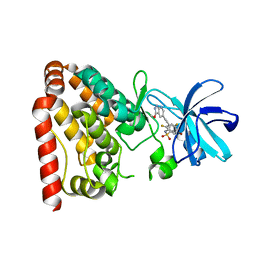 | | Crystal structure of ZAK in complex with compound D2829 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, N-[2,4-bis(fluoranyl)-3-[2-(3-methoxy-1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Dai, Y.B, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure Based Design of N-(3-((1H-Pyrazolo[3,4-b]pyridin-5-yl)ethynyl)benzenesulfonamides as Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J. Med. Chem., 60, 2017
|
|
8FV2
 
 | | Bromodomain of CBP liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FVF
 
 | | Bromodomain of EP300 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Histone acetyltransferase p300, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXE
 
 | | Bromodomain of CBP liganded with iCBP6 | | Descriptor: | (6S)-1-(3-tert-butylphenyl)-6-{(5P)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
7DD1
 
 | |
8FVK
 
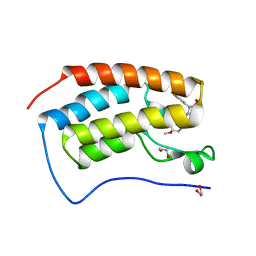 | | First bromodomain of BRD4 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXA
 
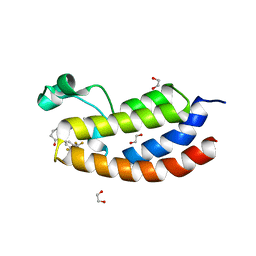 | | Bromodomain of CBP liganded with iCBP4 | | Descriptor: | (6S)-1-[3,5-bis(trifluoromethyl)phenyl]-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXO
 
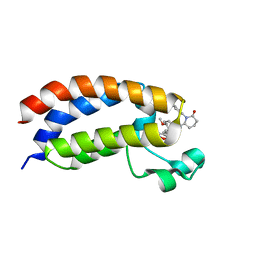 | | Bromodomain of CBP liganded with iCBP8 | | Descriptor: | (6S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | Descriptor: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Fan, S, Wan, W. | | Deposit date: | 2021-04-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ECW
 
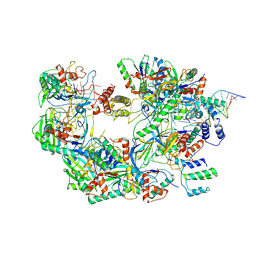 | | The Csy-AcrIF14-dsDNA complex | | Descriptor: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECV
 
 | | The Csy-AcrIF14 complex | | Descriptor: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
4HXX
 
 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
8G6T
 
 | | Bromodomain of CBP liganded with inhibitor iCBP2 | | Descriptor: | (6S)-1-(3,4-dibromophenyl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein, NICKEL (II) ION | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
7D8M
 
 | | Crystal structure of DyP | | Descriptor: | Dye-decolorizing peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | He, C, Jia, R, Wang, T, Li, L.Q. | | Deposit date: | 2020-10-08 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing two important tryptophan residues with completely different roles in a dye-decolorizing peroxidase from Irpex lacteus F17.
Biotechnol Biofuels, 14, 2021
|
|
6J76
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase in Complex with NAP | | Descriptor: | Aldehyde dehydrogenase A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
4LF3
 
 | |
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4LJR
 
 | |
