6KBX
 
 | |
6KBS
 
 | | Crystal structure of yedK in complex with ssDNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*CP*GP*AP*TP*TP*C)-3'), SOS response-associated protein | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6KCQ
 
 | |
6J4O
 
 | |
6B2E
 
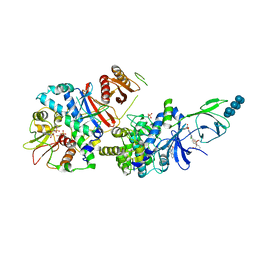 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6B1U
 
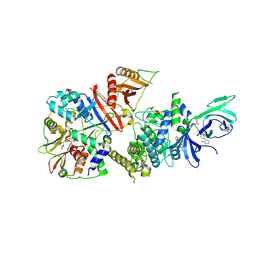 | | Structure of full-length human AMPK (a2b1g1) in complex with a small molecule activator SC4 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
7CIZ
 
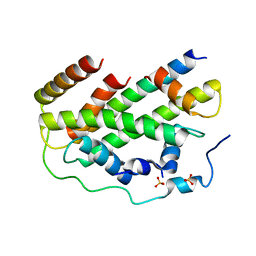 | |
7CJ0
 
 | |
7V1L
 
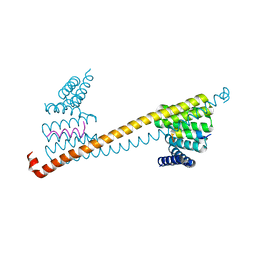 | |
7V1K
 
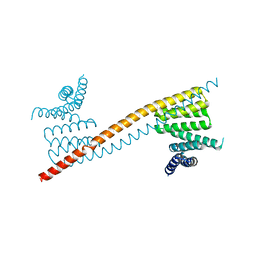 | | Apo structure of sNASP core | | Descriptor: | Isoform 2 of Nuclear autoantigenic sperm protein | | Authors: | Bao, H, Huang, H. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | NASP maintains histone H3-H4 homeostasis through two distinct H3 binding modes.
Nucleic Acids Res., 50, 2022
|
|
7DSY
 
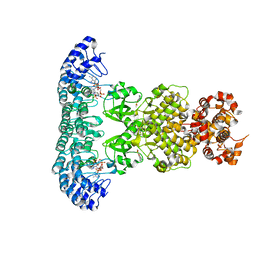 | | Crystal Structure of RNase L in complex with KM05073 | | Descriptor: | 1-chloranyl-3-methylsulfinyl-6,7-dihydro-5H-2-benzothiophen-4-one, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-03 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7DTS
 
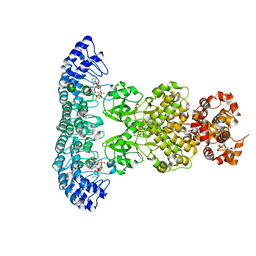 | | Crystal structure of RNase L in complex with AC40357 | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7ELW
 
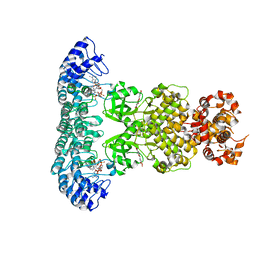 | | Crystal structure of RNase L in complex with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | Descriptor: | UBX domain-containing protein 4 | | Authors: | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | Deposit date: | 2010-05-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
7DG6
 
 | |
7FA3
 
 | |
9JDU
 
 | | The crystal structure of PDE10A complexed with inhibitor 2061 | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2024-09-01 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.3000176 Å) | | Cite: | Discovery of novel azetidine-based imidazopyridines as selective and orally bioavailable inhibitors of phosphodiesterase 10A for the treatment of pulmonary arterial hypertension.
Eur.J.Med.Chem., 290, 2025
|
|
4N4V
 
 | |
4N3R
 
 | | Co-crystal structure of tankyrase 1 with compound 2 (5-(2-aminoquinazolin-6-yl)-N-(4,4-dimethyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)-2-fluorobenzamide) | | Descriptor: | 5-(2-aminoquinazolin-6-yl)-N-(4,4-dimethyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)-2-fluorobenzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-10-07 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of 2-aminopyridine oxazolidinones as potent and selective tankyrase inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4N4T
 
 | |
4HGT
 
 | | Crystal structure of ck1d with compound 13 | | Descriptor: | 2-{2-[(3,4-difluorophenoxy)methyl]-5-methoxypyridin-4-yl}-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform delta | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
4HGL
 
 | | Crystal structure of ck1g3 with compound 1 | | Descriptor: | 2-[5-methoxy-2-(quinolin-3-yl)pyrimidin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
3DPK
 
 | | cFMS tyrosine kinase in complex with a pyridopyrimidinone inhibitor | | Descriptor: | 8-cyclohexyl-N-methoxy-5-oxo-2-{[4-(2-pyrrolidin-1-ylethyl)phenyl]amino}-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, Macrophage colony-stimulating factor 1 receptor, Fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrido[2,3-d]pyrimidin-5-ones: a novel class of antiinflammatory macrophage colony-stimulating factor-1 receptor inhibitors
J.Med.Chem., 52, 2009
|
|
4EUX
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with 6-(5-(((3R,4R)-4-((6-amino-4-methylpyridin-2-yl)methyl)pyrrolidin-3-yl)oxy)pentyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[5-({(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}oxy)pentyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2012-04-25 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Selective monocationic inhibitors of neuronal nitric oxide synthase. Binding mode insights from molecular dynamics simulations.
J.Am.Chem.Soc., 134, 2012
|
|
3BEA
 
 | | cFMS tyrosine kinase (tie2 KID) in complex with a pyrimidinopyridone inhibitor | | Descriptor: | 8-(2,3-dihydro-1H-inden-5-yl)-2-({4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]phenyl}amino)-5-oxo-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schubert, C. | | Deposit date: | 2007-11-16 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Design and synthesis of a pyrido[2,3-d]pyrimidin-5-one class of anti-inflammatory FMS inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
