4WXI
 
 | |
1T17
 
 | | Solution Structure of the 18 kDa Protein CC1736 from Caulobacter crescentus: The Northeast Structural Genomics Consortium Target CcR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Shen, Y, Atreya, H.S, Acton, T, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity.
Proteins, 58, 2005
|
|
6WGH
 
 | |
6QVP
 
 | |
5LAB
 
 | |
4AC7
 
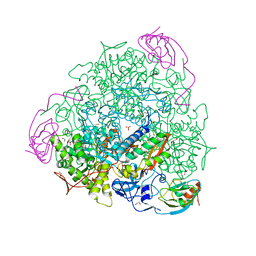 | | The crystal structure of Sporosarcina pasteurii urease in complex with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, HYDROXIDE ION, ... | | Authors: | Benini, S, Kosikowska, P, Cianci, M, Gonzalez Vara, A, Berlicki, L, Ciurli, S. | | Deposit date: | 2011-12-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Sporosarcina Pasteurii Urease in a Complex with Citrate Provides New Hints for Inhibitor Design.
J.Biol.Inorg.Chem., 18, 2013
|
|
2KMU
 
 | |
2K5O
 
 | |
4O7I
 
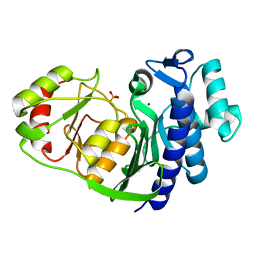 | | Structural and functional characterization of 3'(2'),5'-bisphosphate nucleotidase1 from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Rehman, S.A.A, Gourinath, S. | | Deposit date: | 2013-12-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural elucidation of a dual-activity PAP phosphatase-1 from Entamoeba histolytica capable of hydrolysing both 3'-phosphoadenosine 5'-phosphate and inositol 1,4-bisphosphate
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7Z0Y
 
 | | THSC20.HVTR04 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
7Z0X
 
 | | THSC20.HVTR26 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
2HVQ
 
 | | Structure of Adenylated full-length T4 RNA Ligase 2 | | Descriptor: | Hypothetical 37.6 kDa protein in Gp24-hoc intergenic region, MAGNESIUM ION | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVS
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2JYK
 
 | | NMR Structure of a 21 bp DNA duplex preferentially cleaved by Human Topoisomerase II | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DCP*DAP*DCP*DGP*DT)-3'), DNA (5'-D(*DAP*DCP*DGP*DTP*DGP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DAP*DTP*DAP*DAP*DGP*DCP*DTP*DGP*DT)-3') | | Authors: | Masliah, G, Mauffret, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of intrinsic dynamics in a DNA sequence preferentially cleaved by topoisomerase II enzyme
J.Mol.Biol., 381, 2008
|
|
1MFA
 
 | |
3EB3
 
 | | Voltage-dependent K+ channel beta subunit (W121A) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
8A76
 
 | | Metallo-beta-lactamase NDM-1 in complex with 1,2,4-Triazole-3-thione compound 26 | | Descriptor: | (2~{S})-2-[bis(pyridin-2-ylmethyl)amino]-5-(3-phenyl-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl)pentanoic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Legru, A, Hernandez, J.F, Cendron, L. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of 1,2,4-Triazole-3-thiones toward Broad-Spectrum Metallo-beta-lactamase Inhibitors Showing Potent Synergistic Activity on VIM- and NDM-1-Producing Clinical Isolates.
J.Med.Chem., 65, 2022
|
|
3EB4
 
 | | Voltage-dependent K+ channel beta subunit (I211R) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
2WXD
 
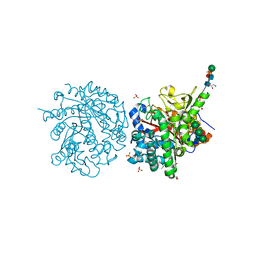 | | A MICROMOLAR O-SULFATED THIOHYDROXIMATE INHIBITOR BOUND TO PLANT MYROSINASE | | Descriptor: | 2-(DIMETHYLAMINO)ETHYL (1Z)-2-PHENYL-N-(SULFOOXY)ETHANIMIDOTHIOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Besle, A, Burmeister, W.P. | | Deposit date: | 2009-11-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Micromolar O-Sulfated Thiohydroximate Inhibitor Bound to Plant Myrosinase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6SKY
 
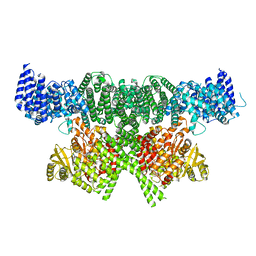 | | FAT and kinase domain of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SL0
 
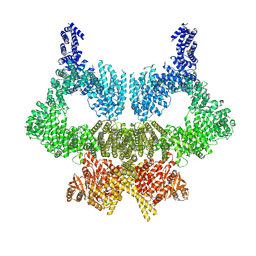 | | Complete CtTel1 dimer with C2 symmetry | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
4ICC
 
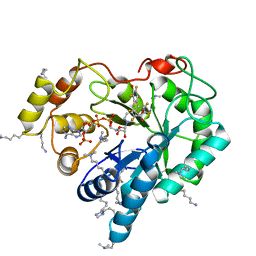 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8GBX
 
 | |
8GBY
 
 | | Crystal structure of PC39-50E, an anti-HIV broadly neutralizing antibody | | Descriptor: | GLYCEROL, PC39-50E Fab heavy chain, PC39-50E Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
