7DSZ
 
 | | Crystal structure of Amuc_1102 from Akkermansia muciniphila | | Descriptor: | Amuc_1102, CHLORIDE ION | | Authors: | Xiang, R, Wang, J, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Amuc_1102 fromAkkermansia muciniphilaadopts animmunoglobulin-like fold related to archaeal type IV pilus
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7PUQ
 
 | | CARM1 in complex with EML982 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]propyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
5XLT
 
 | | The crystal structure of tubulin in complex with 4'-demethylepipodophyllotoxin | | Descriptor: | (5S,5aR,8aR,9R)-9-(3,5-dimethoxy-4-oxidanyl-phenyl)-5-oxidanyl-5a,6,8a,9-tetrahydro-5H-[2]benzofuro[6,5-f][1,3]benzodioxol-8-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Structure of 4'-demethylepipodophyllotoxin in complex with tubulin provides a rationale for drug design
Biochem. Biophys. Res. Commun., 493, 2017
|
|
7PPY
 
 | | CARM1 in complex with EML709 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PV6
 
 | | CARM1 in complex with EML734 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, TETRAETHYLENE GLYCOL, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PU8
 
 | | CARM1 in complex with EML980 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[N-[2-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PPQ
 
 | | CARM1 in complex with EML736 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PUC
 
 | | CARM1 in complex with EML981 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[(~{E})-3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]prop-2-enyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8WBX
 
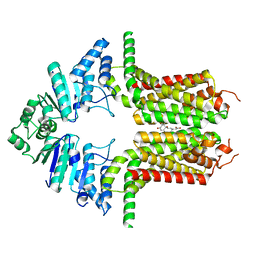 | | Cryo-EM structure of the ABCG25 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-10 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WAM
 
 | |
8WD6
 
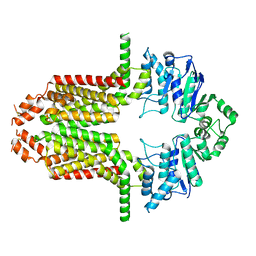 | | Cryo-EM structure of the ABCG25 | | Descriptor: | ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WBA
 
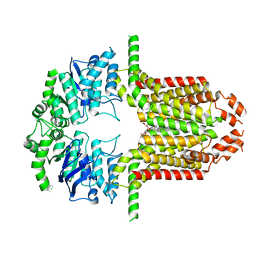 | | Cryo-EM structure of the ABCG25 bound to CHS | | Descriptor: | ABC transporter G family member 25, CHOLESTEROL HEMISUCCINATE | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-09 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC9
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
