5T7I
 
 | | Crystal structure of galectin-8N in complex with LNnT | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
6DU3
 
 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION, REST-pS861 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
6DU2
 
 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | MAGNESIUM ION, REST-pS861/4, carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
3J89
 
 | | Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies | | Descriptor: | synthetic peptide | | Authors: | Egelman, E.H, Xu, C, DiMaio, F, Magnotti, E, Modlin, C, Yu, X, Wright, E, Baker, D, Conticello, V.P. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural plasticity of helical nanotubes based on coiled-coil assemblies.
Structure, 23, 2015
|
|
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
3J9R
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4X4J
 
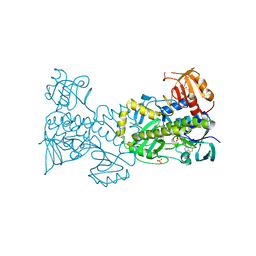 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
2HI5
 
 | | Model for bacteriophage fd from cryo-EM | | Descriptor: | Coat protein B | | Authors: | Wang, Y.A, Yu, X, Overman, S, Tsuboi, M, Thomas, G.J, Egelman, E.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The Structure of a Filamentous Bacteriophage
J.Mol.Biol., 361, 2006
|
|
6O3V
 
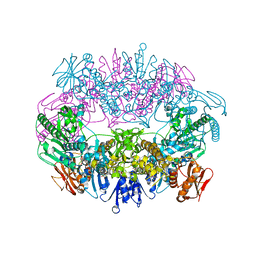 | | Crystal structure for RVA-VP3 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Protein VP3, ... | | Authors: | Kumar, D, Yu, X, Wang, Z, Hu, L, Prasad, V. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2.7 angstrom cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity.
Sci Adv, 6, 2020
|
|
6OVQ
 
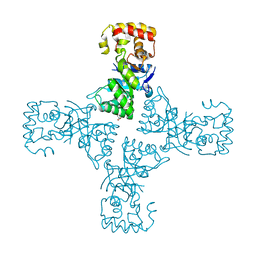 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | Descriptor: | GLYCEROL, Putative Side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5ZCJ
 
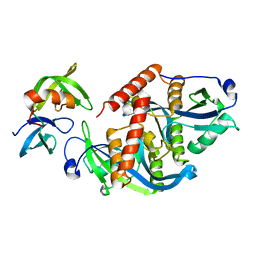 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
4E2I
 
 | | The Complex Structure of the SV40 Helicase Large T Antigen and p68 Subunit of DNA Polymerase Alpha-Primase | | Descriptor: | DNA polymerase alpha subunit B, Large T antigen, ZINC ION | | Authors: | Zhou, B, Arnett, D.R, Yu, X, Brewster, A, Sowd, G.A, Xie, C.L, Vila, S, Gai, D, Fanning, E, Chen, X.S. | | Deposit date: | 2012-03-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase.
J.Biol.Chem., 287, 2012
|
|
7SWD
 
 | | Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound | | Descriptor: | 1C11 scFv, 1C3 heavy chain, 1C3 light chain, ... | | Authors: | Milligan, J.C, Yu, X, Saphire, E.O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Asymmetric and non-stoichiometric glycoprotein recognition by two distinct antibodies results in broad protection against ebolaviruses.
Cell, 185, 2022
|
|
3TXX
 
 | | Crystal structure of putrescine transcarbamylase from Enterococcus faecalis | | Descriptor: | Putrescine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Zhao, G, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-09-23 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of putrescine transcarbamylase from Enterococcus faecalis: Structural insights into the oligomeric assembly and the active site
To be Published
|
|
3S6K
 
 | | Crystal structure of xcNAGS | | Descriptor: | Acetylglutamate kinase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8018 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6G
 
 | | Crystal structures of Seleno-substituted mutant mmNAGS in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, MALONATE ION, ... | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6681 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6H
 
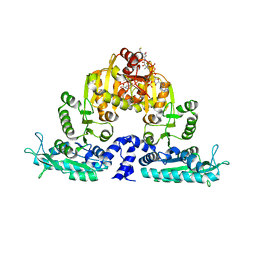 | | Crystal structure of native mmNAGS/k | | Descriptor: | COENZYME A, GLUTAMIC ACID, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
5J8R
 
 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 12 - K61R mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Li, H, Yang, F, Xu, Y.F, Wang, W.J, Xiao, P, Yu, X, Sun, J.P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Crystal structure and substrate specificity of PTPN12.
Cell Rep, 15, 2016
|
|
3S7Y
 
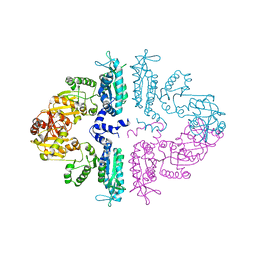 | | Crystal structure of mmNAGS in Space Group P3121 at 4.3 A resolution | | Descriptor: | N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3077 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
4GFU
 
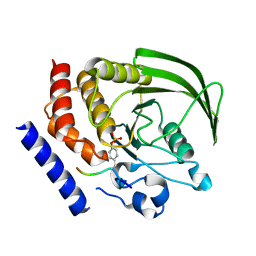 | | PTPN18 in complex with HER2-pY1248 phosphor-peptides | | Descriptor: | HER2-pY1248 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
9MGY
 
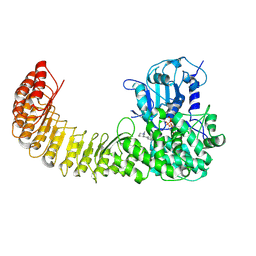 | | Cryo-EM structure of Human NLRP3 complex with compound 1 | | Descriptor: | (2M)-2-(6-{[(3R)-1-methylpiperidin-3-yl]amino}pyridazin-3-yl)-5-(trifluoromethyl)phenol, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Mammoliti, O, Carbajo, R.J, Perez-Benito, L, Yu, X, Prieri, M.L.C, Bontempi, L, Embrechts, S, Paesmans, I, Bassi, M, Bhattacharya, A, Roman, S.C, Hoog, S.D, Demin, S, Gijsen, H.J.M, Hache, G, Jacobs, T, Jerhaoui, S, Leenaerts, J, Lutter, F.H, Matico, R, Oehlrich, D, Perrier, M, Ryabchuk, P, Schepens, W, Sharma, S, Somers, M, Suarez, J, Surkyn, M, Opdenbosch, N.V, Verhulst, T, Bottelbergs, A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Bicyclic NLRP3 Inhibitors with Peripheral and Central In Vivo Activity.
J.Med.Chem., 68, 2025
|
|
9MIG
 
 | | Cryo-EM structure of Human NLRP3 complex with compound 3 | | Descriptor: | (2P)-2-(4-{[(3R)-1-methylpiperidin-3-yl]amino}-5,6,7,8-tetrahydrophthalazin-1-yl)-5-(trifluoromethyl)phenol, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Mammoliti, O, Carbajo, R.J, Perez-Benito, L, Yu, X, Prieri, M.L.C, Bontempi, L, Embrechts, S, Paesmans, I, Bassi, M, Bhattacharya, A, Roman, S.C, Hoog, S.D, Demin, S, Gijsen, H.J.M, Hache, G, Jacobs, T, Jerhaoui, S, Leenaerts, J, Lutter, F.H, Matico, R, Oehlrich, D, Perrier, M, Ryabchuk, P, Schepens, W, Sharma, S, Somers, M, Suarez, J, Surkyn, M, Opdenbosch, N.V, Verhulst, T, Bottelbergs, A. | | Deposit date: | 2024-12-12 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Bicyclic NLRP3 Inhibitors with Peripheral and Central In Vivo Activity.
J.Med.Chem., 68, 2025
|
|
9MIE
 
 | | Human NLRP3 complex with compound 2 in the closed hexamer | | Descriptor: | (2P)-2-(4-{[(3R)-1-methylpiperidin-3-yl]amino}-6,7-dihydro-5H-cyclopenta[d]pyridazin-1-yl)-5-(trifluoromethyl)phenol, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Mammoliti, O, Carbajo, R.J, Perez-Benito, L, Yu, X. | | Deposit date: | 2024-12-12 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Bicyclic NLRP3 Inhibitors with Peripheral and Central In Vivo Activity.
J.Med.Chem., 68, 2025
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
