3FC2
 
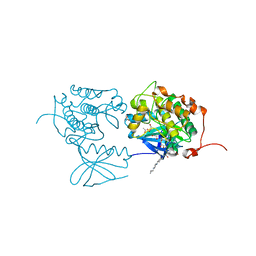 | | PLK1 in complex with BI6727 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bader, G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BI 6727, A Polo-like Kinase Inhibitor with Improved Pharmacokinetic Profile and Broad Antitumor Activity.
Clin.Cancer Res., 15, 2009
|
|
1ODU
 
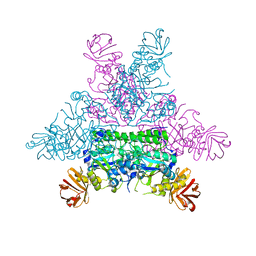 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH FUCOSE | | Descriptor: | PUTATIVE ALPHA-L-FUCOSIDASE, beta-L-fucopyranose | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
3CUI
 
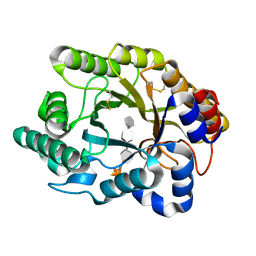 | |
6EK8
 
 | | YaxB from Yersinia enterocolitica | | Descriptor: | YaxB | | Authors: | Braeuning, B, Groll, M. | | Deposit date: | 2017-09-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
3GVA
 
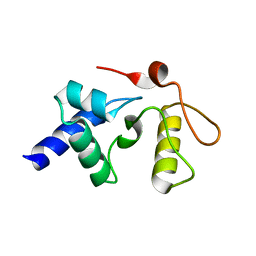 | |
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
3ST3
 
 | | Dreiklang - off state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3ST2
 
 | | Dreiklang - equilibrium state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3ST4
 
 | | Dreiklang - on state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3BO8
 
 | | The High Resolution Crystal Structure of HLA-A1 Complexed with the MAGE-A1 Peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational changes within the HLA-A1:MAGE-A1 complex induced by binding of a recombinant antibody fragment with TCR-like specificity
Protein Sci., 18, 2009
|
|
7PH1
 
 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
7T0O
 
 | |
7T0R
 
 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | Authors: | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
4QXS
 
 | | Crystal structure of human FPPS in complex with WC01088 | | Descriptor: | (2-{2-[(2S)-3-methylbutan-2-yl]-5-phenyl-1H-indol-3-yl}ethane-1,1-diyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Park, J, Zielinski, M, Weiling, C, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the molecular and structural elements of ligands binding to the active site versus an allosteric pocket of the human farnesyl pyrophosphate synthase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1AUW
 
 | | H91N DELTA 2 CRYSTALLIN FROM DUCK | | Descriptor: | DELTA 2 CRYSTALLIN | | Authors: | Abu-Abed, M, Vallee, F, Howell, P.L. | | Deposit date: | 1997-09-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of the enzymatically active and inactive forms of delta crystallin and the role of histidine 91.
Biochemistry, 36, 1997
|
|
1OD8
 
 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine lactam | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, SODIUM ION, ... | | Authors: | Gloster, T.M, Roberts, S, Davies, G.J. | | Deposit date: | 2003-02-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Xylobiose-Derived Isofagomine Lactam Glycosidase Inhibitor Binds as its Amide Tautomer
Chem.Commun.(Camb.), 8, 2003
|
|
2Q2A
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ARGININE, ArtJ, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-26 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2POX
 
 | |
2Q2C
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, GLYCEROL, HISTIDINE, ... | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2PVU
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, LYSINE, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2QYP
 
 | |
2R0R
 
 | |
2RB3
 
 | | Crystal Structure of Human Saposin D | | Descriptor: | GLYCEROL, Proactivator polypeptide, SULFATE ION | | Authors: | Maier, T, Rossman, M, Saenger, W. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human saposins C and d: implications for lipid recognition and membrane interactions.
Structure, 16, 2008
|
|
2R1Q
 
 | |
1NDA
 
 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
