7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
7VAD
 
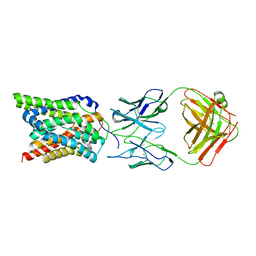 | | Cryo-EM structure of human NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Sodium/bile acid cotransporter | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7VAE
 
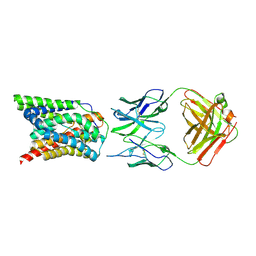 | | Cryo-EM structure of bovine NTCP complexed with YN69202Fab | | Descriptor: | Fab heavy chain from antibody IgG clone number YN69202, Fab light chain from antibody IgG clone number YN69202, Solute carrier family 10 (Sodium/bile acid cotransporter family), ... | | Authors: | Asami, J, Shimizu, T, Ohto, U. | | Deposit date: | 2021-08-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the bile acid transporter and HBV receptor NTCP.
Nature, 606, 2022
|
|
7U5K
 
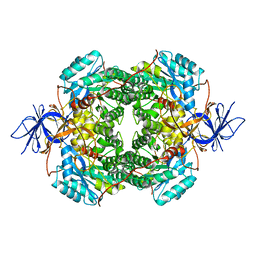 | | Cryo-EM Structure of DPYSL2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7TLX
 
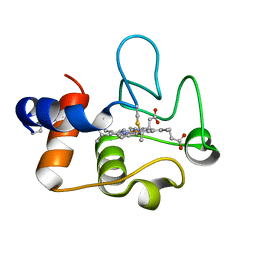 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
7VTQ
 
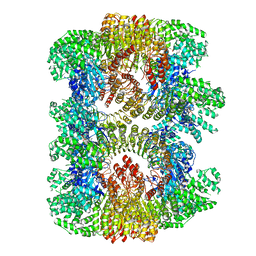 | | Cryo-EM structure of mouse NLRP3 (full-length) dodecamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VTP
 
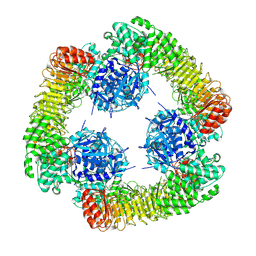 | | Cryo-EM structure of PYD-deleted human NLRP3 hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGI
 
 | |
7VGH
 
 | |
7VGJ
 
 | | Cryo-EM structure of the human P4-type flippase ATP8B1-CDC50A in the auto-inhibited E2Pi-PS state | | Descriptor: | Cell cycle control protein 50A, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Phospholipid-transporting ATPase IC | | Authors: | Chen, M.T, Chen, Y, Chen, Z.P, Zhou, C.Z, Hou, W.T, Chen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural insights into the activation of autoinhibited human lipid flippase ATP8B1 upon substrate binding.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8ZBQ
 
 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
6L56
 
 | | Fe(II) loaded Tegillarca granosa ferritin | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85300577 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
4BXF
 
 | |
7WRY
 
 | |
6L4Z
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 6 | | Descriptor: | 4-(hydroxymethyl)benzoic acid, Genome polyprotein | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
6L55
 
 | | Recombinant Tegillarca granosa ferritin | | Descriptor: | FE (III) ION, FORMIC ACID, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78304863 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
6KZY
 
 | | Cu(II) loaded Tegillarca granosa ferritin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-09-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.30057073 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
6L50
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | Descriptor: | 2-sulfanylidene-1,3-thiazolidin-4-one, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
6L58
 
 | | Cu(II) loaded Tegillarca granosa M-ferritin soaked with Fe(II) | | Descriptor: | COPPER (II) ION, Ferritin | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.90270972 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
7DMO
 
 | |
5Y81
 
 | | NuA4 TEEAA sub-complex | | Descriptor: | Actin, Actin-related protein 4, Chromatin modification-related protein EAF1, ... | | Authors: | Wang, X, Cai, G. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex
Nat Commun, 9, 2018
|
|
5W99
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis bound to TSP | | Descriptor: | 2,2'-(6-(2'-(aminomethyl)-[2,4'-bithiazol]-4-yl)pyridine-2,5-diyl)bis(thiazole-4-carboxylic acid), PbtD, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7EIL
 
 | | BRD4-BD1 in complex with LT-909-110 | | Descriptor: | Bromodomain-containing protein 4, N-[4-[4-ethanoyl-5-methyl-2-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-pyrrol-3-yl]phenyl]ethanamide | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 1-(5-(1H-benzo[d]imidazole-2-yl)-2,4-dimethyl-1H-pyrrol-3-yl)ethan-1-one derivatives as novel and potent bromodomain and extra-terminal (BET) inhibitors with anticancer efficacy
Eur.J.Med.Chem., 227, 2022
|
|
5WA4
 
 | |
6MN6
 
 | |
