2IO5
 
 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
8OU1
 
 | |
4X5X
 
 | |
4X5W
 
 | | HLA-DR1 with CLIP102-120(M107W) | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Guenther, S, Freund, C. | | Deposit date: | 2014-12-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | MHC class II complexes sample intermediate states along the peptide exchange pathway.
Nat Commun, 7, 2016
|
|
6AAJ
 
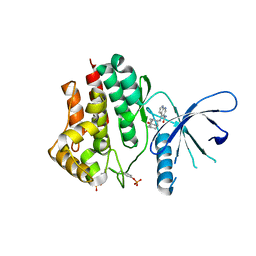 | | Crystal structure of JAK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAM
 
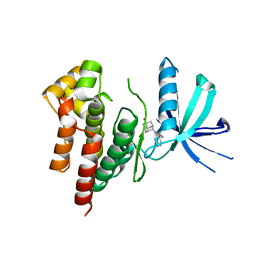 | | Crystal structure of TYK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Nomura, N, Tomimoto, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAH
 
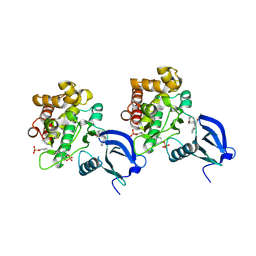 | | Crystal structure of JAK1 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAK
 
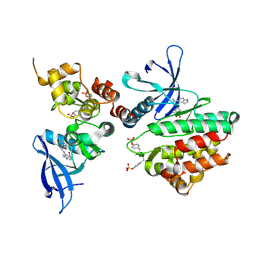 | | Crystal structure of JAK3 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
4YN8
 
 | | Crystal Structure of Response Regulator ChrA in Heme-Sensing Two Component System | | Descriptor: | MAGNESIUM ION, Response regulator ChrA, SULFATE ION | | Authors: | Doi, A, Nakamura, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2015-03-09 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3DOM
 
 | | Crystal Structure of the complex between Tfb5 and the C-terminal domain of Tfb2 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, RNA polymerase II transcription factor B subunit 5 | | Authors: | Kainov, D.E, Cavarelli, J, Egly, J.M, Poterszman, A. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for group A trichothiodystrophy
Nat.Struct.Mol.Biol., 15, 2008
|
|
3KHW
 
 | |
3L56
 
 | | Crystal structure of the large c-terminal domain of polymerase basic protein 2 from influenza virus a/viet nam/1203/2004 (h5n1) | | Descriptor: | Polymerase PB2 | | Authors: | Staker, B.L, Edwards, T, Eric, S, Raymond, A, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biological and structural characterization of a host-adapting amino acid in influenza virus.
Plos Pathog., 6, 2010
|
|
3KC6
 
 | |
5URM
 
 | | Crystal structure of human BRR2 in complex with T-1206548 | | Descriptor: | 3-(5-{[(2R)-5-amino-2-cyclohexyl-7-oxo-2,3-dihydro-7H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-6-yl]methyl}furan-2-yl)benzoic acid, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URJ
 
 | | Crystal structure of human BRR2 in complex with T-3905516 | | Descriptor: | 6-benzyl-3-[(2R)-2-(3-fluoropyridin-2-yl)-6-methyl-3,4-dihydro-2H-1-benzopyran-7-yl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(3H,8H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URK
 
 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
1JDO
 
 | |
7X8W
 
 | |
7X91
 
 | |
7X8Z
 
 | |
7X8Y
 
 | |
7X90
 
 | |
7X92
 
 | |
1LOP
 
 | | CYCLOPHILIN A COMPLEXED WITH SUCCINYL-ALA-PRO-ALA-P-NITROANILIDE | | Descriptor: | CYCLOPHILIN A, SUCCINYL-ALA-PRO-ALA-P-NITROANILIDE | | Authors: | Konno, M. | | Deposit date: | 1996-06-17 | | Release date: | 1996-12-23 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The substrate-binding site in Escherichia coli cyclophilin A preferably recognizes a cis-proline isomer or a highly distorted form of the trans isomer.
J.Mol.Biol., 256, 1996
|
|
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
