4JAN
 
 | | crystal structure of broadly neutralizing antibody CH103 in complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
5U78
 
 | | Crystal structure of ORP8 PH domain in P1211 space group | | Descriptor: | Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
6DS5
 
 | | Cryo EM structure of human SEIPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Seipin | | Authors: | Yan, R.H, Qian, H.W, Yan, N, Yang, H.Y. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human SEIPIN Binds Anionic Phospholipids.
Dev. Cell, 47, 2018
|
|
4N79
 
 | | Structure of Cathepsin K-dermatan sulfate complex | | Descriptor: | Cathepsin K, alpha-L-idopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of collagen fiber degradation by cathepsin K.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JAM
 
 | | Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 | | Descriptor: | 1,2-ETHANEDIOL, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
8FYG
 
 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8FNX
 
 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8G0O
 
 | | Crystal structure of Y281F mutant of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
7VAH
 
 | |
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp4/5 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp14/15 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E5X
 
 | |
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
5ZM8
 
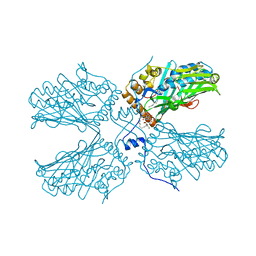 | | Crystal structure of ORP2-ORD in complex with PI(4,5)P2 | | Descriptor: | Oxysterol-binding protein-related protein 2, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | Authors: | Wang, H, Dong, J.Q, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ORP2 Delivers Cholesterol to the Plasma Membrane in Exchange for Phosphatidylinositol 4, 5-Bisphosphate (PI(4,5)P2).
Mol. Cell, 73, 2019
|
|
8HOK
 
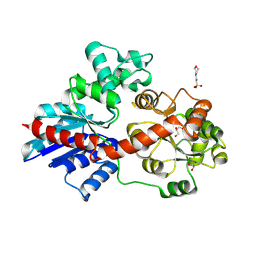 | | crystal structure of UGT71AP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, UGT71AP2 | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
8HOJ
 
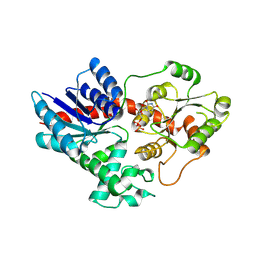 | | Crystal structure of UGT71AP2 in complex with UDP | | Descriptor: | UGT71AP2, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
8HIJ
 
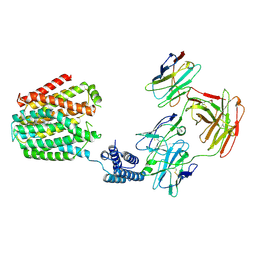 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
7ZCZ
 
 | |
