5ZZ2
 
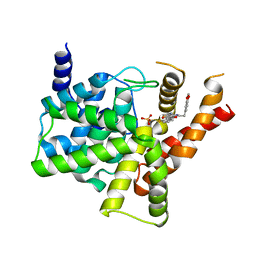 | | Crystal structure of PDE5 in complex with inhibitor LW1634 | | Descriptor: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
6ACB
 
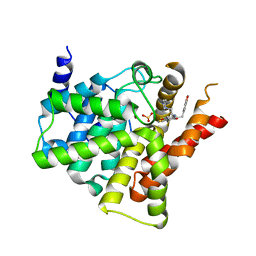 | | Crystal structure of PDE5 in complex with inhibitor LW1805 | | Descriptor: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-6-{[2-(4-methylpiperazin-1-yl)ethyl]amino}-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2018-07-26 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
7XT2
 
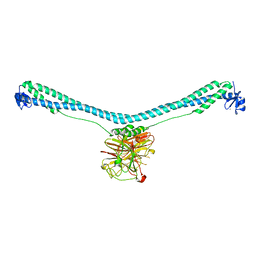 | | Crystal structure of TRIM72 | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Zhou, C, Ma, Y.M, Ding, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for TRIM72 oligomerization during membrane damage repair.
Nat Commun, 14, 2023
|
|
7XP1
 
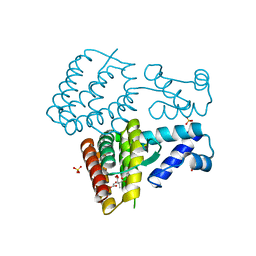 | | Crystal structure of PmiR from Pseudomonas aeruginosa | | Descriptor: | ALPHA-METHYLISOCITRIC ACID, GLYCEROL, Probable transcriptional regulator, ... | | Authors: | Zhang, Y.X, Liang, H.H, Gan, J.H. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa.
Sci Adv, 8, 2022
|
|
6B77
 
 | | Structures of the two-chain human plasma factor XIIa co-crystallized with potent inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL, ... | | Authors: | Dementiev, A.A, Silva, A, Yee, C, Flavin, M.T, Partridge, J.R. | | Deposit date: | 2017-10-03 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of human plasma beta-factor XIIa cocrystallized with potent inhibitors.
Blood Adv, 2, 2018
|
|
6BFP
 
 | | Bovine trypsin bound to potent inhibitor | | Descriptor: | 3-{2-[(4-carbamimidoylphenyl)carbamoyl]-4-ethenyl-5-methoxyphenyl}-6-[(cyclopropylmethyl)carbamoyl]pyridine-2-carboxylic acid, CALCIUM ION, Cationic trypsin | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2017-10-26 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
6B74
 
 | |
8FWD
 
 | |
8GME
 
 | | Crystal structure of the gp32-Dda-dT17 complex | | Descriptor: | Dda helicase, ZINC ION, dT17, ... | | Authors: | He, X, Yun, M.K, White, S.W. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-28 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (4.98 Å) | | Cite: | Structural and functional insights into the interaction between the bacteriophage T4 DNA processing proteins gp32 and Dda.
Nucleic Acids Res., 52, 2024
|
|
8GOF
 
 | | Structure of hSLC19A1+PMX | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8F2B
 
 | | Amylin 3 Receptor in complex with Gs and Pramlintide analogue peptide San45 | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Sexton, P.M, Wootten, D.L, Belousoff, M.J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural insight into selectivity of amylin and calcitonin receptor agonists.
Nat.Chem.Biol., 20, 2024
|
|
8F0K
 
 | | Human Amylin3 Receptor in complex with Gs and Pramlintide analogue peptide San385 | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Sexton, P.M, Wootten, D.L. | | Deposit date: | 2022-11-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structural insight into selectivity of amylin and calcitonin receptor agonists.
Nat.Chem.Biol., 20, 2024
|
|
8G9J
 
 | |
8G9K
 
 | |
8GA6
 
 | |
8GA7
 
 | |
8F0J
 
 | | Calcitonin Receptor in complex with Gs and Pramlintide analogue peptide San45 | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Sexton, P.M, Wootten, D.L, Belousoff, M.J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural insight into selectivity of amylin and calcitonin receptor agonists.
Nat.Chem.Biol., 20, 2024
|
|
8F2A
 
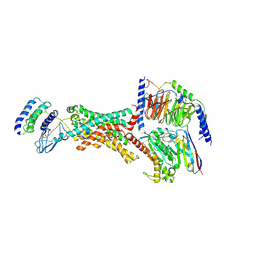 | | Human Amylin3 Receptor in complex with Gs and Pramlintide analogue peptide San385 (Cluster 5 conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, J, Sexton, P.M, Wootten, D.L, Radostin, D. | | Deposit date: | 2022-11-07 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural insight into selectivity of amylin and calcitonin receptor agonists.
Nat.Chem.Biol., 20, 2024
|
|
8G4K
 
 | |
8GAC
 
 | | Crystal structure of a high affinity CTLA-4 binder | | Descriptor: | 1,2-ETHANEDIOL, CTLA-4 binder | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
