7CH4
 
 | |
7CHC
 
 | |
7CH5
 
 | |
4FHZ
 
 | | Crystal structure of a carboxyl esterase at 2.0 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phospholipase/Carboxylesterase, SODIUM ION | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
7CKK
 
 | | Structural complex of FTO bound with Dac51 | | Descriptor: | 2-{[2,6-dichloro-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino}-N-hydroxybenzamide, Alpha-ketoglutarate-dependent dioxygenase FTO, N-OXALYLGLYCINE | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance.
Cell Metab., 33, 2021
|
|
4NT6
 
 | | HLA-C*0801 Crystal Structure | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, Cw-8 alpha chain, ... | | Authors: | Liu, J.X, Toh, X.Y, Ren, E.C. | | Deposit date: | 2013-12-01 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans.
J.Virol., 88, 2014
|
|
4IU6
 
 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | Descriptor: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | Deposit date: | 2013-01-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
2JXT
 
 | | Solution structure of 50S ribosomal protein LX from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium (NESG) target TR80 | | Descriptor: | 50S ribosomal protein LX | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein LX from Methanobacterium thermoautotrophicum.
To be Published
|
|
2JN0
 
 | | Solution NMR structure of the ygdR protein from Escherichia coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Hypothetical lipoprotein ygdR | | Authors: | Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-15 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ygdR protein from Escherichia coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
8GJY
 
 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJX
 
 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJZ
 
 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
5GH9
 
 | | Crystal structure of CBP Bromodomain with H3K56ac peptide | | Descriptor: | CREB-binding protein, Histone H3 | | Authors: | Xu, L. | | Deposit date: | 2016-06-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insight into CBP bromodomain-mediated recognition of acetylated histone H3K56ac
FEBS J., 2017
|
|
7Y3G
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | G-protein coupled receptor 12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, H, Zhang, J.Y, Luo, F. | | Deposit date: | 2022-06-10 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insight into the constitutive activity of human orphan receptor GPR12.
Sci Bull (Beijing), 68, 2023
|
|
6O1F
 
 | |
2JWY
 
 | | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli. Northeast Structural Genomics target ER540 | | Descriptor: | Uncharacterized lipoprotein yajI | | Authors: | Liu, G, Xiao, R, Chen, C, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli.
To be Published
|
|
4E4K
 
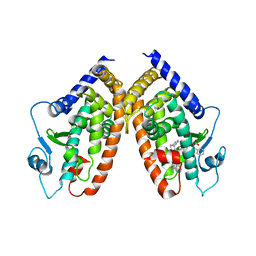 | | Crystal Structure of PPARgamma with the ligand JO21 | | Descriptor: | (2S)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4E4Q
 
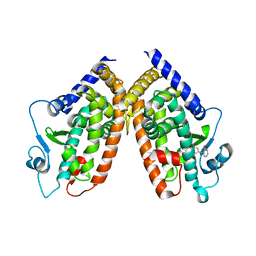 | | Crystal structure of PPARgamma with the ligand FS214 | | Descriptor: | (2R)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
7Y99
 
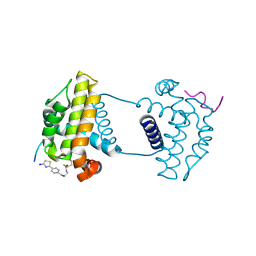 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YAA
 
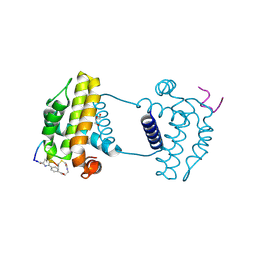 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y90
 
 | | Crystal Structure Analysis of cp1 bound BCL2 | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W. | | Deposit date: | 2022-06-24 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YA5
 
 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YB7
 
 | | anti-apoptotic protein BCL-2-M12 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide, cp2 peptide | | Authors: | Li, F.W, Liu, C, Wu, D.L. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
5ZNG
 
 | | The crystal complex of immune receptor RGA5A_S of Pia from rice (Oryzae sativa) with rice blast (Magnaporthe oryzae) effector protein AVR1-CO39 | | Descriptor: | AVR1-CO39, NBS-LRR type protein | | Authors: | Guo, L.W, Zhang, Y.K, Liu, Q, Ma, M.Q, Liu, J.F, Peng, Y.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
