8I5E
 
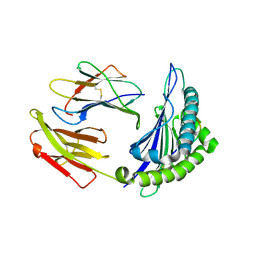 | | Crystal structure of HLA-A*11:01 in complex with KRAS peptide (VVGAGGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12wt-9, MHC class I antigen (Fragment) | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
6J4R
 
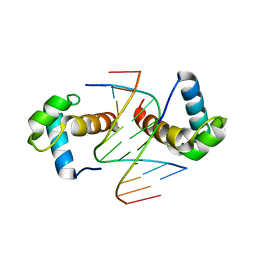 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6J4K
 
 | |
6J5B
 
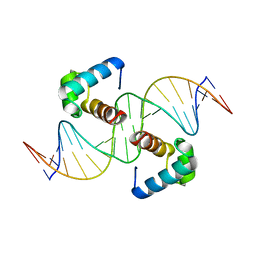 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6RCI
 
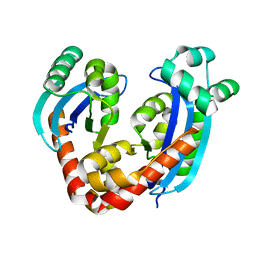 | | Crystal structure of REXO2 | | Descriptor: | Oligoribonuclease, mitochondrial | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6RCN
 
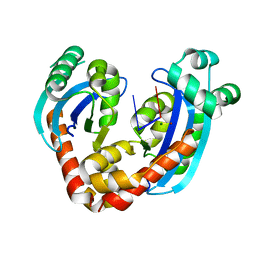 | | Crystal structure of REXO2-D199A-dAdA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), MAGNESIUM ION, Oligoribonuclease, ... | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6RCL
 
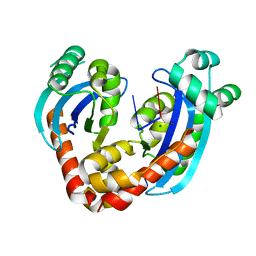 | | Crystal structure of REXO2-D199A-AA | | Descriptor: | MAGNESIUM ION, Oligoribonuclease, mitochondrial, ... | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6P5G
 
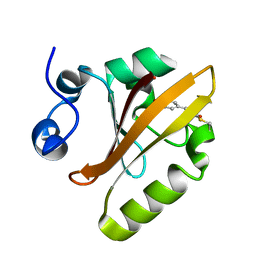 | | Photoactive Yellow Protein PYP Dark Full | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P4I
 
 | | Photoactive Yellow Protein PYP 10ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5E
 
 | | Photoactive Yellow Protein PYP 80ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
6P5D
 
 | | Photoactive Yellow Protein PYP 30ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
4EML
 
 | |
4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | Descriptor: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | Descriptor: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | Authors: | Meng, R, Chang, J, Zhang, J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
3HSS
 
 | | A higher resolution structure of Rv0554 from Mycobacterium tuberculosis complexed with malonic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3HYS
 
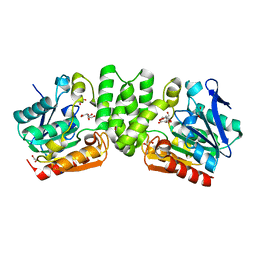 | | Structure of Rv0554 from Mycobacterium tuberculosis complexed with Malonic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MALONIC ACID, ... | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2009-06-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4BU2
 
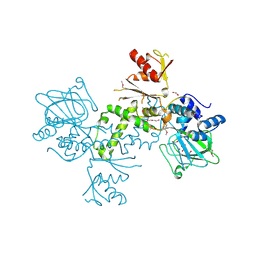 | | 60S ribosomal protein L27A histidine hydroxylase (MINA53) in complex with Ni(II) and 2-oxoglutarate (2OG) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE MINA, ... | | Authors: | Chowdhury, R, Clifton, I.J, McDonough, M.A, Ng, S.S, Pilka, E, Oppermann, U, Schofield, C.J. | | Deposit date: | 2013-06-19 | | Release date: | 2014-05-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
3E3A
 
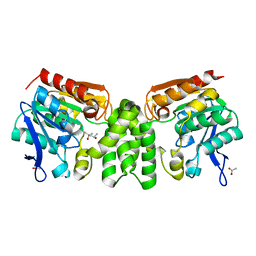 | | The Structure of Rv0554 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, POSSIBLE PEROXIDASE BPOC | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2008-08-06 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4CCK
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II) and N-oxalylglycine (NOG) | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCN
 
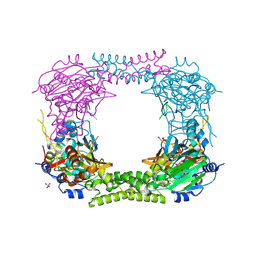 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 L299C/C300S) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-2) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
