7BRQ
 
 | | Crystal structure of human FAM134B LIR fused to human GABARAP | | Descriptor: | GLYCEROL, Reticulophagy regulator 1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRT
 
 | | Crystal structure of Sec62 LIR fused to GABARAP | | Descriptor: | Translocation protein SEC62,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
3PBP
 
 | |
3X26
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 5 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X25
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 700 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X24
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 120 min | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X20
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 25 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, N, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7BQ3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ4
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA)-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5X9O
 
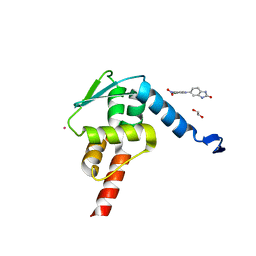 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X9P
 
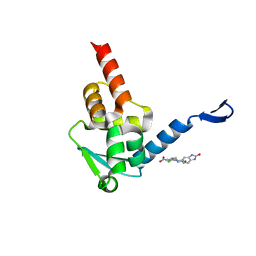 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
6OIM
 
 | |
7ZZX
 
 | |
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP5
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome (Structure ii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
1IQQ
 
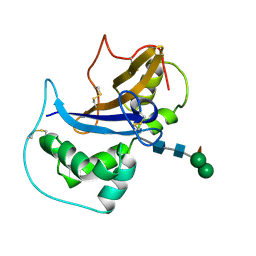 | | Crystal Structure of Japanese pear S3-RNase | | Descriptor: | S3-RNase, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Matsuura, T, Sakai, H, Norioka, S. | | Deposit date: | 2001-07-25 | | Release date: | 2001-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility.
J.Biol.Chem., 276, 2001
|
|
8WU5
 
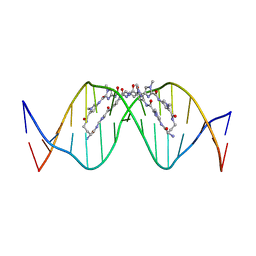 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | Descriptor: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | Authors: | Abe, K, Takeda, K, Sugiyama, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
7NS0
 
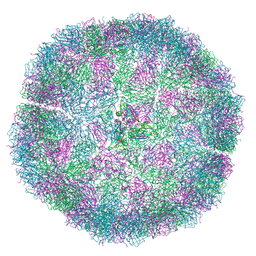 | | Bacilladnavirus capsid structure | | Descriptor: | Capsid protein VP2 | | Authors: | Munke, A, Okamoto, K. | | Deposit date: | 2021-03-05 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses.
Mbio, 13, 2022
|
|
2YW9
 
 | |
8ABX
 
 | | Crystal structure of IDO1 in complex with Apoxidole-1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, O1-tert-butyl O2-ethyl O5-methyl (E,5R)-5-(1-methylindol-2-yl)-5-[(4-methylphenyl)sulfonylamino]pent-2-ene-1,2,5-tricarboxylate, O2-tert-butyl O3-ethyl O6-methyl (2S,6R)-6-(1-methylindol-2-yl)-2,5-dihydro-1H-pyridine-2,3,6-tricarboxylate, ... | | Authors: | Dotsch, L, Ziegler, S, Waldmann, H, Gasper, R. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Pseudo-Natural Product Type IV IDO1 Inhibitor Chemotype.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7VSG
 
 | |
7VSH
 
 | |
7W47
 
 | | Crystal structure of the gastric proton pump complexed with tegoprazan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Potassium-transporting ATPase alpha chain 1, ... | | Authors: | Abe, K, Tanaka, S, Morita, M, Yamagishi, T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump.
J.Med.Chem., 65, 2022
|
|
7W48
 
 | |
7W49
 
 | | Crystal structure of the gastric proton pump complexed with soraprazan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Potassium-transporting ATPase alpha chain 1, ... | | Authors: | Abe, K, Tanaka, S. | | Deposit date: | 2021-11-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump.
J.Med.Chem., 65, 2022
|
|
