6P4G
 
 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5N
 
 | | Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1 | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5K
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
3IZ4
 
 | |
4C4Q
 
 | | Cryo-EM map of the CSFV IRES in complex with the small ribosomal 40S subunit and DHX29 | | Descriptor: | INTERNAL RIBOSOMAL ENTRY SITE | | Authors: | Hashem, Y, desGeorges, A, Dhote, V, Langlois, R, Liao, H.Y, Grassucci, R.A, Pestova, T.V, Hellen, C.U.T, Frank, J. | | Deposit date: | 2013-09-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Hepatitis-C-Virus-Like Internal Ribosome Entry Sites Displace Eif3 to Gain Access to the 40S Subunit
Nature, 503, 2013
|
|
4V7H
 
 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
2BCW
 
 | | Coordinates of the N-terminal domain of ribosomal protein L11,C-terminal domain of ribosomal protein L7/L12 and a portion of the G' domain of elongation factor G, as fitted into cryo-em map of an Escherichia coli 70S*EF-G*GDP*fusidic acid complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L7/L12, Elongation factor G | | Authors: | Datta, P.P, Sharma, M.R, Qi, L, Frank, J, Agrawal, R.K. | | Deposit date: | 2005-10-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Interaction of the G' Domain of Elongation Factor G and the C-Terminal Domain of Ribosomal Protein L7/L12 during Translocation as Revealed by Cryo-EM.
Mol.Cell, 20, 2005
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
7SYH
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
2P8Z
 
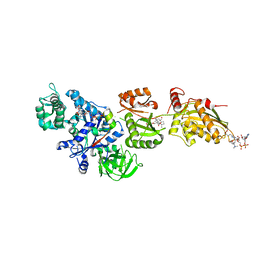 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8W
 
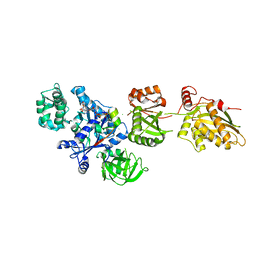 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
1LU3
 
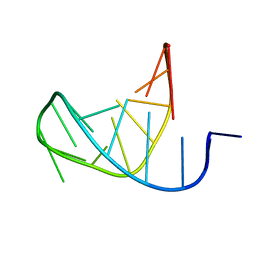 | | Separate Fitting of the Anticodon Loop Region of tRNA (nucleotide 26-42) in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | PHENYLALANINE TRANSFER RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
1LS2
 
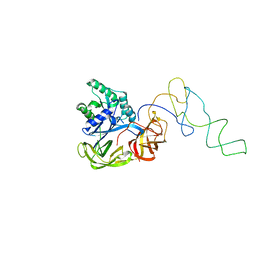 | | Fitting of EF-Tu and tRNA in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | Elongation Factor Tu, Phenylalanine transfer RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-16 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
2P8X
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8Y
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
6OUO
 
 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
2FVO
 
 | | Docking of the modified RF1 X-ray structure into the Low Resolution Cryo-EM map of E.coli 70S Ribosome bound with RF1 | | Descriptor: | Peptide chain release factor 1 | | Authors: | Rawat, U, Gao, H, Zavialov, A, Gursky, R, Ehrenberg, M, Frank, J. | | Deposit date: | 2006-01-31 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Interactions of the Release Factor RF1 with the Ribosome as Revealed by Cryo-EM.
J.Mol.Biol., 357, 2006
|
|
3JBO
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P/E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
3JBP
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBN
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
