2A4Z
 
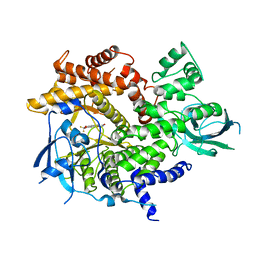 | | Crystal Structure of human PI3Kgamma complexed with AS604850 | | Descriptor: | (5E)-5-[(2,2-DIFLUORO-1,3-BENZODIOXOL-5-YL)METHYLENE]-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-06-30 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
2A5U
 
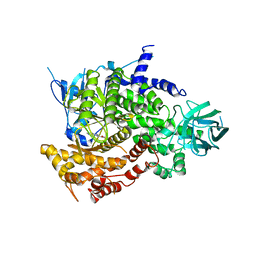 | | Crystal Structure of human PI3Kgamma complexed with AS605240 | | Descriptor: | (5E)-5-(QUINOXALIN-6-YLMETHYLENE)-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
6NVR
 
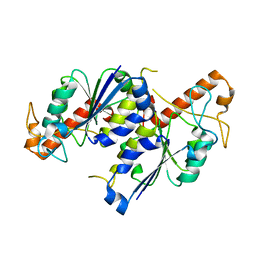 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in Apo form | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V.M, Blundell, T.L. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.562 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6NW7
 
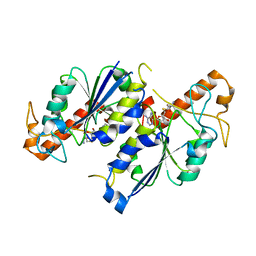 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6NW6
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with S-Adenosyl methionine | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6TW9
 
 | | HumRadA22F in complex with CAM833 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW3
 
 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | Descriptor: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
5L3B
 
 | | Human LSD1/CoREST: LSD1 D556G mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5L3C
 
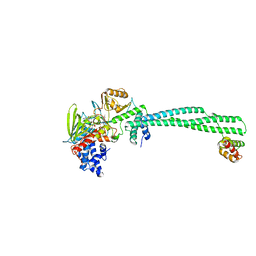 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5LB2
 
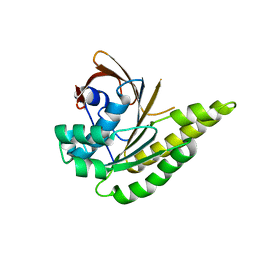 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5MWO
 
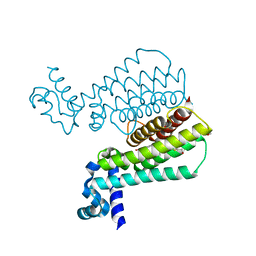 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1ZYZ
 
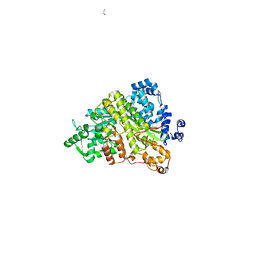 | | Structures of Yeast Ribonucloetide Reductase I | | Descriptor: | GLYCINE, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5MXK
 
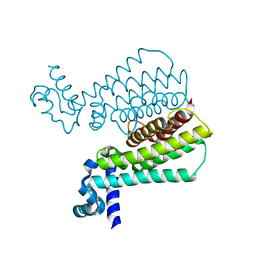 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7G9. | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1ZZD
 
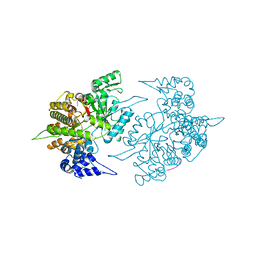 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | Ribonucleoside-diphosphate reductase large chain 1, Ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2X7P
 
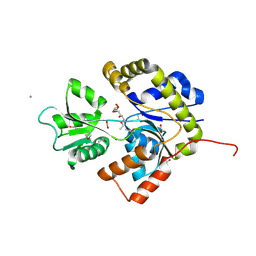 | | The Conserved Candida albicans CA3427 Gene Product Defines a New Family of Proteins Exhibiting the Generic Periplasmic Binding Protein Structural Fold | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Santini, S, Monchois, V, Mouz, N, Rousselle, T, Claverie, J.M, Abergel, C. | | Deposit date: | 2010-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Conserved Candida Albicans Ca3427 Gene Product Defines a New Family of Proteins Exhibiting the Generic Periplasmic Binding Protein Structural Fold
Plos One, 6, 2011
|
|
2X7Q
 
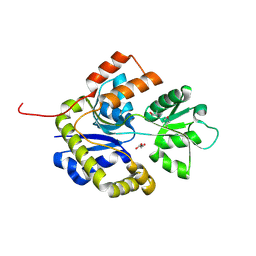 | | The conserved Candida albicans CA3427 gene product defines a new family of proteins exhibiting the generic periplasmic binding protein structural fold | | Descriptor: | CALCIUM ION, GLYCEROL, POSSIBLE THIAMINE BIOSYNTHESIS ENZYME | | Authors: | Santini, S, Monchois, V, Mouz, N, Rousselle, T, Claverie, J.M, Abergel, C. | | Deposit date: | 2010-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Conserved Candida Albicans Ca3427 Gene Product Defines a New Family of Proteins Exhibiting the Generic Periplasmic Binding Protein Structural Fold
Plos One, 6, 2011
|
|
3KIP
 
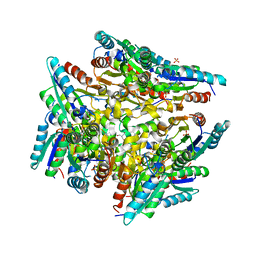 | | Crystal structure of type-II 3-dehydroquinase from C. albicans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinase, type II, ... | | Authors: | Trapani, S, Schoehn, G, Navaza, J, Abergel, C. | | Deposit date: | 2009-11-02 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7PTV
 
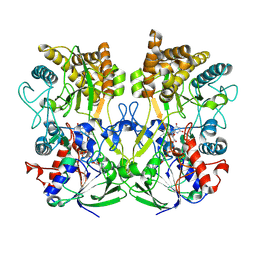 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7QRJ
 
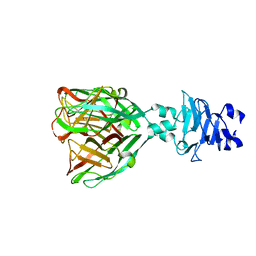 | | Crystal structure of Zamilon vitis protein Zav_19 | | Descriptor: | Zav_19 protein | | Authors: | Jeudy, S, Abergel, C. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The fibre head structure used by unrelated families of viruses is unexpectedly a major component of the Marseilleviridae and Zamilon virophages capsids
To Be Published
|
|
7QRR
 
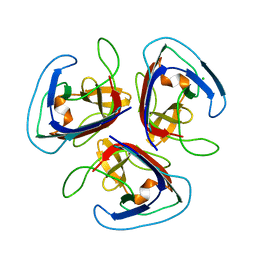 | | Crystal structure of Noumeavirus NMV_189 protein | | Descriptor: | CHLORIDE ION, NMV_189 protein, PHOSPHATE ION | | Authors: | Jeudy, S, Abergel, C. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The fibre head structure used by unrelated families of viruses is unexpectedly a major component of the Marseilleviridae and Zamilon virophages capsids
To Be Published
|
|
6YY6
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-(1-ethylpyrazol-4-yl)pyrimidine-5-carbonitrile, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
6YYC
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-6-[1-[(3-fluorophenyl)methyl]pyrazol-4-yl]pyrimidin-4-amine, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
6YVQ
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in apo form | | Descriptor: | 1,2-ETHANEDIOL, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
6YX3
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
