6BTZ
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group C121 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
6BTY
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group P41212 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
5NHR
 
 | |
1Z30
 
 | | NMR structure of the apical part of stemloop D from cloverleaf 1 of bovine enterovirus 1 RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*CP*GP*UP*UP*AP*GP*AP*AP*CP*GP*UP*C)-3' | | Authors: | Ihle, Y, Ohlenschlager, O, Duchardt, E, Ramachandran, R, Gorlach, M. | | Deposit date: | 2005-03-10 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA
Nucleic Acids Res., 33, 2005
|
|
5NGV
 
 | |
5CTN
 
 | | Structure of BPu1 beta-lactamase | | Descriptor: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-25 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
6BU0
 
 | | Crystal structure of the PI3KC2alpha C2 domain in complex with IP6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, FORMIC ACID, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
6BUB
 
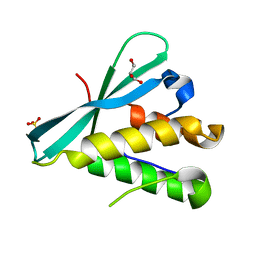 | | Crystal structure of the PI3KC2alpha PX domain in space group P432 | | Descriptor: | GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
3N26
 
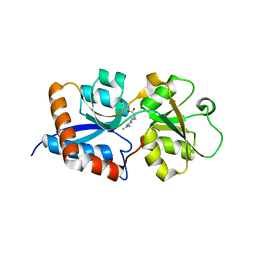 | | Cpn0482 : the arginine binding protein from the periplasm of chlamydia Pneumoniae | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Petit, P, Garcia, C, Vuillard, L, Soriani, M, Grandi, G, Marseilles Structural Genomics Program AFMB (MSGP), Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting antigenic diversity for vaccine design: the Chlamydia ArtJ paradigm.
J.Biol.Chem., 2010
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
5HIA
 
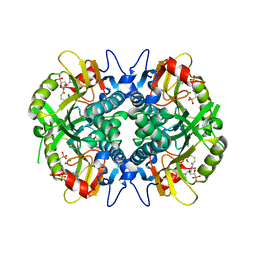 | | Human hypoxanthine-guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Guddat, L.W, Keough, D.T, Rejman, D. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 2017
|
|
6HKA
 
 | |
1YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH HISTIDINE | | Descriptor: | HISTIDINE, HYDROXYNITRILE LYASE, SULFATE ION | | Authors: | Wagner, U.G, Kratky, C. | | Deposit date: | 1996-05-15 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of cyanogenesis: the crystal structure of hydroxynitrile lyase from Hevea brasiliensis.
Structure, 4, 1996
|
|
7JTZ
 
 | | Yeast Glo3 GAP domain | | Descriptor: | ADP-ribosylation factor GTPase-activating protein GLO3, GLYCEROL, ZINC ION | | Authors: | Xie, B, Jackson, L.P. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Glo3 GAP crystal structure supports the molecular niche model for ArfGAPs in COPI coats.
Adv Biol Regul, 79, 2021
|
|
3MAV
 
 | | Crystal structure of Plasmodium vivax putative farnesyl pyrophosphate synthase (Pv092040) | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION | | Authors: | Dong, A, Dunford, J, Lew, J, Wernimont, A.K, Ren, H, Zhao, Y, Koeieradzki, I, Opperman, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
4QLB
 
 | | Structural Basis for the Recruitment of Glycogen Synthase by Glycogenin | | Descriptor: | GLYCEROL, Probable glycogen [starch] synthase, Protein GYG-1, ... | | Authors: | Zeqiraj, E, Judd, A, Sicheri, F. | | Deposit date: | 2014-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recruitment of glycogen synthase by glycogenin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZXH
 
 | |
2LT2
 
 | |
4JLG
 
 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BLB
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL9
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLA
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YAS
 
 | |
1QJ4
 
 | |
3G1Q
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in ligand free state | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha-demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Harp, J, Wawrzak, Z, Waterman, M.R, Park, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections.
J.Biol.Chem., 285, 2010
|
|
