1AAF
 
 | | NUCLEOCAPSID ZINC FINGERS DETECTED IN RETROVIRUSES: EXAFS STUDIES ON INTACT VIRUSES AND THE SOLUTION-STATE STRUCTURE OF THE NUCLEOCAPSID PROTEIN FROM HIV-1 | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Henderson, L.E, Chance, M.R, Bess Junior, J.W, South, T.L, Blake, P.R, Sagi, I, Perez-Alvarado, G, Sowder, R.C, Hare, D.R, Arthur, L.O. | | Deposit date: | 1992-04-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1.
Protein Sci., 1, 1992
|
|
1OMI
 
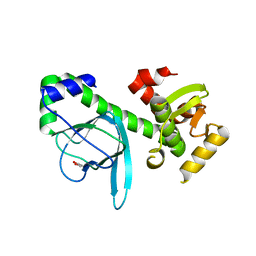 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
3K9A
 
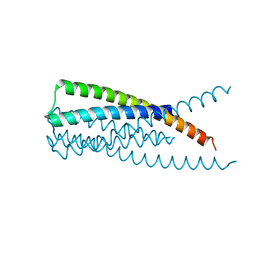 | | Crystal Structure of HIV gp41 with MPER | | Descriptor: | HIV glycoprotein gp41 | | Authors: | Shi, W, Han, D, Habte, H, Cho, M, Chance, M.R. | | Deposit date: | 2009-10-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HIV gp41 with the membrane-proximal external region
J.Biol.Chem., 285, 2010
|
|
2AJT
 
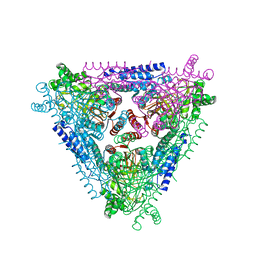 | | Crystal structure of L-Arabinose Isomerase from E.coli | | Descriptor: | L-arabinose isomerase | | Authors: | Manjasetty, B.A, Fedorov, E.V, Almo, S.C, Chance, M.R, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli L-Arabinose Isomerase (ECAI), The Putative Target of Biological Tagatose Production
J.Mol.Biol., 360, 2006
|
|
2XR7
 
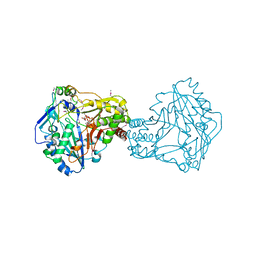 | | Crystal Structure of Nicotiana tabacum malonyltransferase (NtMat1) complexed with malonyl-coa | | Descriptor: | MALONYL-COENZYME A, MALONYLTRANSFERASE | | Authors: | Manjasetty, B.A, Yu, X.H, Panjikar, S, Taguchi, G, Chance, M.R, Liu, C.J. | | Deposit date: | 2010-09-11 | | Release date: | 2011-09-21 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Modification of Flavonol and Naphthol Glucoconjugates by Nicotiana Tabacum Malonyltransferase (Ntmat1).
Planta, 236, 2012
|
|
4F2D
 
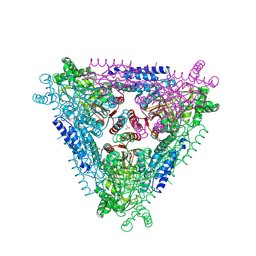 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | Descriptor: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | Authors: | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
1L9G
 
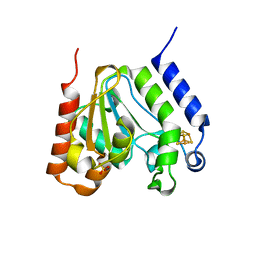 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
3TVI
 
 | | Crystal structure of Clostridium acetobutylicum aspartate kinase (CaAK): An important allosteric enzyme for industrial amino acids production | | Descriptor: | ASPARTIC ACID, Aspartokinase, LYSINE | | Authors: | Manjasetty, B.A, Chance, M.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-09-20 | | Release date: | 2011-11-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Clostridium acetobutylicum Aspartate kinase (CaAK): An important allosteric enzyme for amino acids production.
Biotechnol Rep (Amst), 3, 2014
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
