8Q76
 
 | |
8Q73
 
 | |
7CWS
 
 | | SARS-CoV-2 Spike Proteins Trimer in Complex with FC05 and H014 Fabs Cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of FC05 Fab, ... | | Authors: | Wang, L, Wang, X. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7XFR
 
 | |
7NUE
 
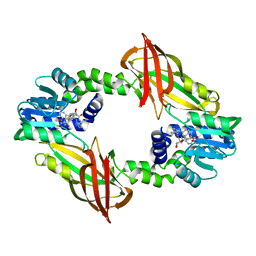 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7NUD
 
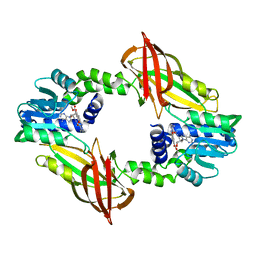 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7XGW
 
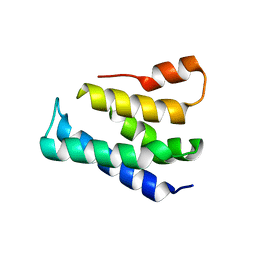 | |
7XY9
 
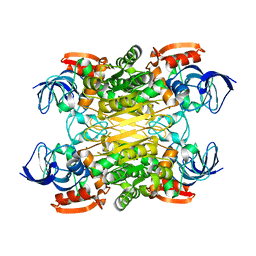 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
6V2R
 
 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V3N
 
 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V8W
 
 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5YOV
 
 | | Crystal structure of BRD4-BD1 bound with hjp126 | | Descriptor: | (3~{R})-4-cyclopentyl-~{N}-(2,4-dimethylphenyl)-1,3-dimethyl-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Hu, J, Li, Y, Cao, D. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure-based optimization of a series of selective BET inhibitors containing aniline or indoline groups.
Eur.J.Med.Chem., 150, 2018
|
|
9CDD
 
 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Laser-Flash-Illuminated | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
9CDC
 
 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Dark State | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium channelrhodopsin 1 C110A, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
9CDE
 
 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Continuous Illumination State | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1 C110A - Continuous Illumination State, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
8E59
 
 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of Amiodarone at 3.1 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E5B
 
 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of Amiodarone and Sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E5A
 
 | | Human L-type voltage-gated calcium channel Cav1.3 treated with 1.4 mM Sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
