7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1WTL
 
 | |
1NRS
 
 | |
1MJZ
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D97N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJY
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D70N | | Descriptor: | INORGANIC PYROPHOSPHATASE | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJW
 
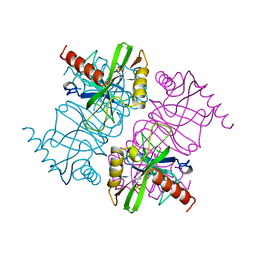 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D42N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Samygina, V.R, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1MJX
 
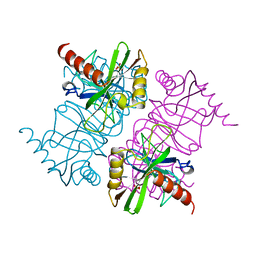 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D65N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
1NRQ
 
 | |
1NRP
 
 | |
1NRN
 
 | |
1NZR
 
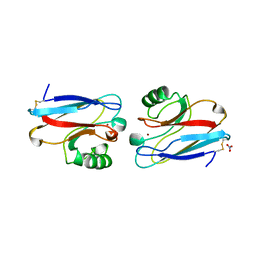 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1NRR
 
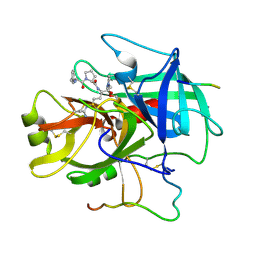 | |
1NRO
 
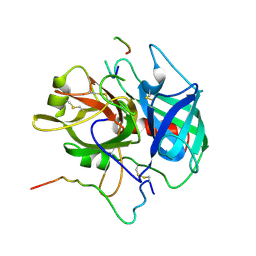 | |
1NN6
 
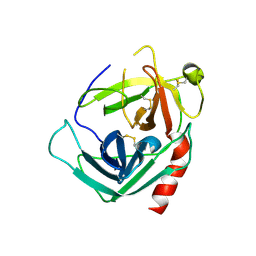 | | Human Pro-Chymase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase | | Authors: | Reiling, K.K, Krucinski, J, Miercke, L.J.W, Raymond, W.W, Caughey, G.H, Stroud, R.M. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human pro-chymase: a model for the activating transition of granule-associated proteases.
Biochemistry, 42, 2003
|
|
1OGY
 
 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | Authors: | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | Deposit date: | 2003-05-19 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
1HA7
 
 | | STRUCTURE OF A LIGHT-HARVESTING PHYCOBILIPROTEIN, C-PHYCOCYANIN FROM SPIRULINA PLATENSIS AT 2.2A RESOLUTION | | Descriptor: | C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, PHYCOCYANOBILIN | | Authors: | Padyana, A.K, Rajashankar, K.R, Ramakumar, S. | | Deposit date: | 2001-03-29 | | Release date: | 2002-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Light-Harvesting Protein C-Phycocyanin from Spirulina Platensis
Biochem.Biophys.Res.Commun., 282, 2001
|
|
1GSF
 
 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
1HXF
 
 | |
1HAG
 
 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUGEN, PRETHROMBIN 2 | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
1GSD
 
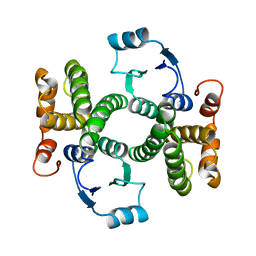 | |
1GSE
 
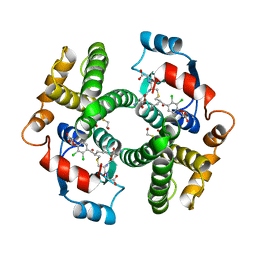 | |
5FV5
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11 | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
1HAI
 
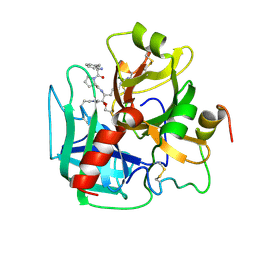 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
1HAH
 
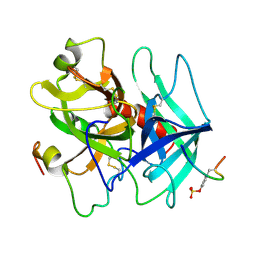 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
1HXE
 
 | | SERINE PROTEASE | | Descriptor: | HIRUDIN VARIANT-1, RUBIDIUM ION, THROMBIN | | Authors: | Tulinsky, A, Zhang, E. | | Deposit date: | 1995-12-07 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular environment of the Na+ binding site of thrombin.
Biophys.Chem., 63, 1997
|
|
