4Q2T
 
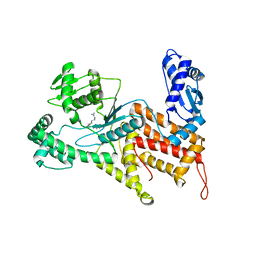 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-arginine | | Descriptor: | ARGININE, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
4Q2X
 
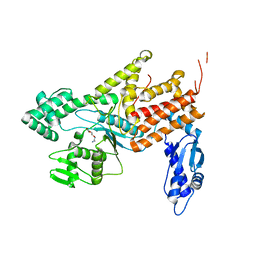 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-canavanine | | Descriptor: | Arginine--tRNA ligase, cytoplasmic, L-CANAVANINE | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
4TR1
 
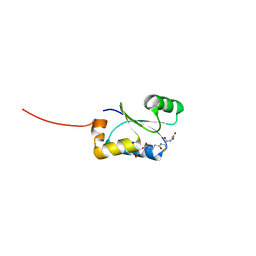 | | Crystal structure of GSH-bound cGrx2/C15S | | Descriptor: | GLUTATHIONE, Glutaredoxin 3 | | Authors: | Lee, E.H, Hwang, K.Y. | | Deposit date: | 2014-06-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | The GSH- and GSSG-bound structures of glutaredoxin from Clostridium oremlandii.
Arch.Biochem.Biophys., 564C, 2014
|
|
4TR0
 
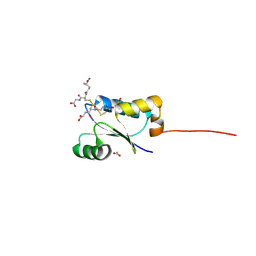 | | Crystal structure of GSSG-bound cGrx2 | | Descriptor: | ACETATE ION, Glutaredoxin 3, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Lee, E.H, Hwang, K.Y. | | Deposit date: | 2014-06-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The GSH- and GSSG-bound structures of glutaredoxin from Clostridium oremlandii.
Arch.Biochem.Biophys., 564C, 2014
|
|
5CHI
 
 | | Crystal structure of PF2046 in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), MAGNESIUM ION, Uncharacterized protein | | Authors: | Kim, J.S, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural basis of two-nucleotide removal of ssDNA by a cryptic RNase H fold 3'-5' exonuclease PF2046 from Pyrococcus furiosus
to be published
|
|
5CZY
 
 | | Crystal structure of LegAS4 | | Descriptor: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | Authors: | Son, J, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
3B6R
 
 | | Crystal structure of Human Brain-type Creatine Kinase | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, ... | | Authors: | Bong, S.M, Moon, J.H, Hwang, K.Y. | | Deposit date: | 2007-10-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
5ZG4
 
 | | Crystal Structure of Triosephosphate isomerase SAD deletion mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
6AA7
 
 | |
4OH9
 
 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OH8
 
 | | Crystal Structure of the human MST1-RASSF5 SARAH heterodimer | | Descriptor: | Ras association domain-containing protein 5, Serine/threonine-protein kinase 4 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1UDT
 
 | | Crystal structure of Human Phosphodiesterase 5 complexed with Sildenafil(Viagra) | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
1UHO
 
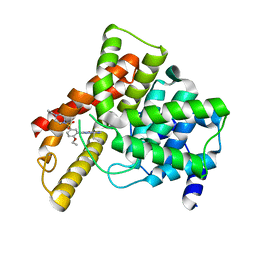 | | Crystal structure of Human Phosphodiesterase 5 complexed with Vardenafil(Levitra) | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-07-09 | | Release date: | 2004-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
1UKH
 
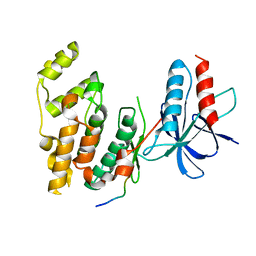 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | Descriptor: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, Mitogen-activated protein kinase 8 isoform 4 | | Authors: | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | Deposit date: | 2003-08-23 | | Release date: | 2004-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
1UKI
 
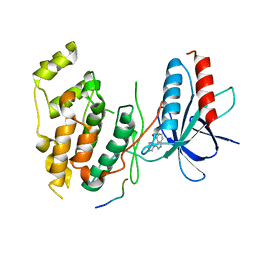 | | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125 | | Descriptor: | 11-mer peptide from C-jun-amino-terminal kinase interacting protein 1, 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, mitogen-activated protein kinase 8 isoform 4 | | Authors: | Heo, Y.-S, Kim, Y.K, Sung, B.-J, Lee, H.S, Lee, J.I, Seo, C.I, Park, S.-Y, Kim, J.H, Hyun, Y.-L, Jeon, Y.H, Ro, S, Lee, T.G, Cho, J.M, Hwang, K.Y, Yang, C.-H. | | Deposit date: | 2003-08-23 | | Release date: | 2004-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Embo J., 23, 2004
|
|
1UDU
 
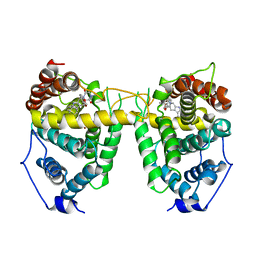 | | Crystal structure of Human Phosphodiesterase 5 complexed with tadalafil(Cialis) | | Descriptor: | 6-BENZO[1,3]DIOXOL-5-YL-2-METHYL-2,3,6,7,12,12A-HEXAHYDRO-PYRAZINO[1',2':1,6]PYRIDO[3,4-B]INDOLE-1,4-DIONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
1IZ2
 
 | | Interactions causing the kinetic trap in serpin protein folding | | Descriptor: | alpha-D-glucopyranose-(1-2)-(5R)-5-[(2R)-2-hydroxynonyl]-beta-D-xylulofuranose, alpha1-antitrypsin | | Authors: | Im, H, Woo, M.-S, Hwang, K.Y, Yu, M.-H. | | Deposit date: | 2002-09-19 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions causing the kinetic trap in serpin protein folding
J.BIOL.CHEM., 277, 2002
|
|
1L8L
 
 | | Molecular basis for the local confomational rearrangement of human phosphoserine phosphatase | | Descriptor: | D-2-AMINO-3-PHOSPHONO-PROPIONIC ACID, L-3-phosphoserine phosphatase | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase.
J.Biol.Chem., 277, 2002
|
|
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
6KIE
 
 | | Crystal structure of human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-07-18 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
6KID
 
 | | Crystal structure of human leucyl-tRNA synthetase, ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-07-18 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
3OIC
 
 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (apo form) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], SULFATE ION | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
6KQY
 
 | | Crystal structure of human leucyl-tRNA synthetase, Leucine-bound form | | Descriptor: | LEUCINE, Leucine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
6KR7
 
 | | Crystal structure of methylated human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-08-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
3OIG
 
 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and INH) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
