2Y42
 
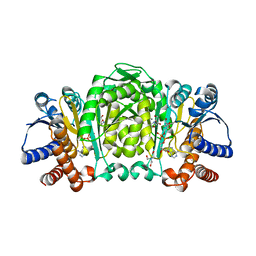 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with NADH and Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, BICINE, MANGANESE (II) ION, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
3C75
 
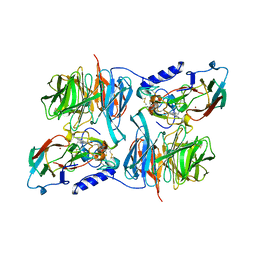 | | Paracoccus versutus methylamine dehydrogenase in complex with amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, Methylamine dehydrogenase heavy chain, ... | | Authors: | Cavalieri, C, Biermann, N, Vlasie, M.D, Einsle, O, Merli, A, Ferrari, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of crystal and solution states of the 138 kDa complex of methylamine dehydrogenase and amicyanin from Paracoccus versutus.
Biochemistry, 47, 2008
|
|
2XE6
 
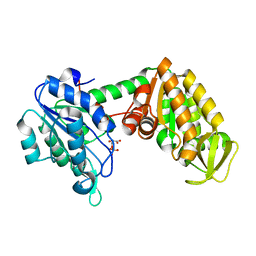 | | The complete reaction cycle of human phosphoglycerate kinase: The open binary complex with 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XE8
 
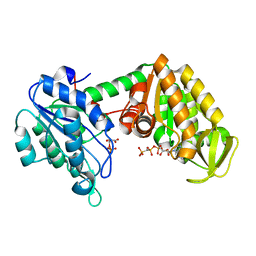 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and AMP-PNP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XE7
 
 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
1JVL
 
 | | Azurin dimer, covalently crosslinked through bis-maleimidomethylether | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
2H47
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 1) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
1JVO
 
 | | Azurin dimer, crosslinked via disulfide bridge | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
2H3X
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes Faecalis (Form 3) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
1HET
 
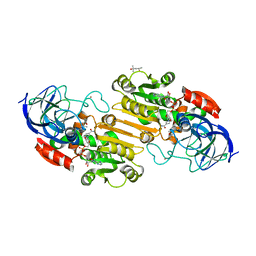 | | atomic X-ray structure of liver alcohol dehydrogenase containing a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-25 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HEU
 
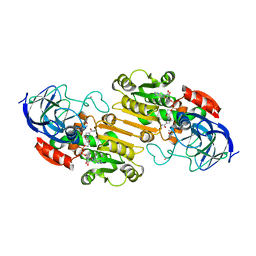 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HF3
 
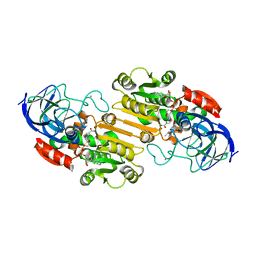 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
3D1K
 
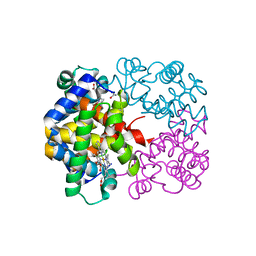 | | R/T intermediate quaternary structure of an antarctic fish hemoglobin in an alpha(CO)-beta(pentacoordinate) state | | Descriptor: | ACETYL GROUP, CARBON MONOXIDE, Hemoglobin subunit alpha-1, ... | | Authors: | Vitagliano, L, Vergara, A, Bonomi, G, Merlino, A, Mazzarella, L. | | Deposit date: | 2008-05-06 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopic and crystallographic characterization of a tetrameric hemoglobin oxidation reveals structural features of the functional intermediate relaxed/tense state.
J.Am.Chem.Soc., 130, 2008
|
|
8RI5
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 48 h of soaking | | Descriptor: | AMMONIA, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.415 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8RI3
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 7 days of soaking | | Descriptor: | AMMONIA, CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
4LFX
 
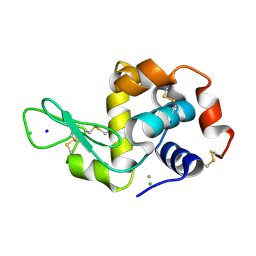 | |
4LGK
 
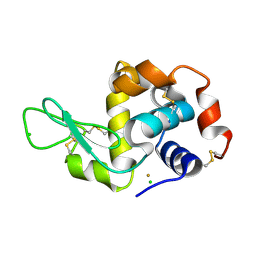 | |
4LFP
 
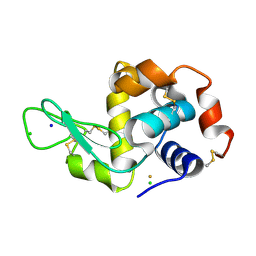 | |
1TQ9
 
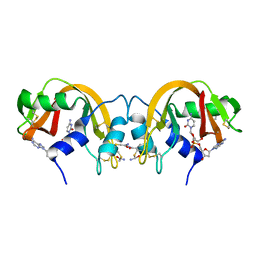 | | Non-covalent swapped dimer of Bovine Seminal Ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Sica, F, Di Fiore, A, Merlino, A, Mazzarella, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Stability of the Non-covalent Swapped Dimer of Bovine Seminal Ribonuclease: AN ENZYME TAILORED TO EVADE RIBONUCLEASE PROTEIN INHIBITOR
J.Biol.Chem., 279, 2004
|
|
5JLG
 
 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and compound I, a piano-stool organometallic Ru(II) arene compound containing an O,S-chelating ligand | | Descriptor: | DIMETHYL SULFOXIDE, RUTHENIUM ION, Ribonuclease pancreatic, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(ii) piano-stool compound containing an O,S-chelating ligand.
Dalton Trans, 45, 2016
|
|
5JVX
 
 | |
8B7L
 
 | |
8B7O
 
 | |
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-11 | | Release date: | 1999-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
4DII
 
 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of potassium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
