6N38
 
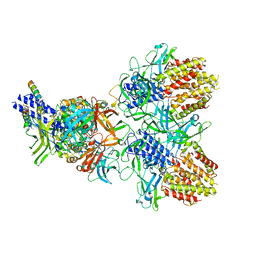 | | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy - full map sharpened | | Descriptor: | Putative type VI secretion protein, Unassigned protein | | Authors: | Park, Y.J, Lacourse, K.D, Cambillau, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy.
Nat Commun, 9, 2018
|
|
6N46
 
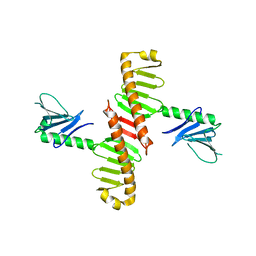 | |
6OG2
 
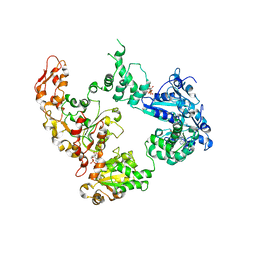 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAX
 
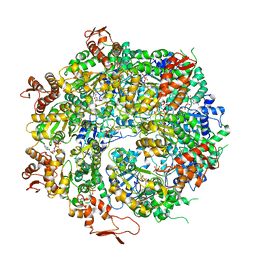 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6DRV
 
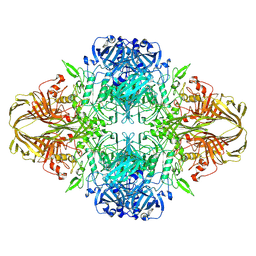 | |
6MZJ
 
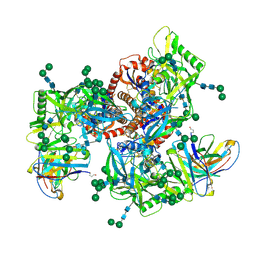 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6OR6
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (tail region) | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6OG3
 
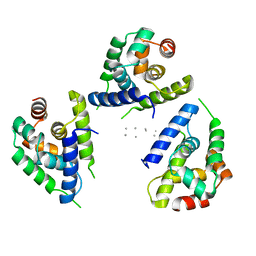 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, NTD-trimer | | Descriptor: | Alpha S1-casein, Hyperactive disaggregase ClpB | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6BQM
 
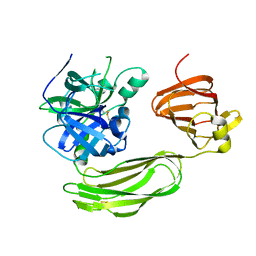 | | Secreted serine protease VesC from Vibrio cholerae | | Descriptor: | serine protease VesC | | Authors: | Park, Y.J, Korotkov, K.V, Delarosa, J.R, Turley, S, DiMaio, F, Hol, W.G.J. | | Deposit date: | 2017-11-28 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Suppressor Mutations in Type II Secretion Mutants of Vibrio cholerae: Inactivation of the VesC Protease.
Msphere, 5, 2020
|
|
6Z5U
 
 | |
4UOT
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE 5H2L | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
4UOS
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
3ZYT
 
 | |
8DT0
 
 | |
3J1W
 
 | |
6HY0
 
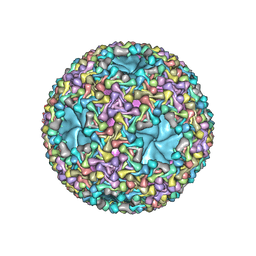 | | Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | | Descriptor: | Major Outer Capsid Protein P8, Major inner protein P1, Packaging Enzyme P4 | | Authors: | El Omari, K, Ilca, S.L, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2018-10-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus.
Nature, 570, 2019
|
|
6Q04
 
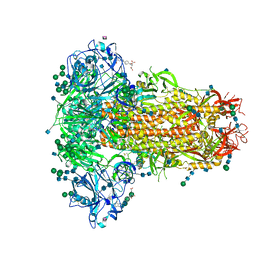 | | MERS-CoV S structure in complex with 5-N-acetyl neuraminic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q06
 
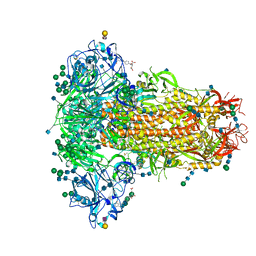 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q05
 
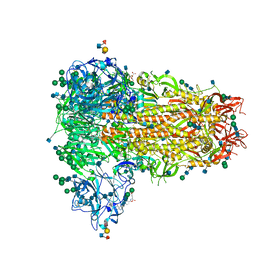 | | MERS-CoV S structure in complex with sialyl-lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q07
 
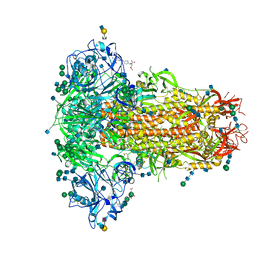 | | MERS-CoV S structure in complex with 2,6-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6MFT
 
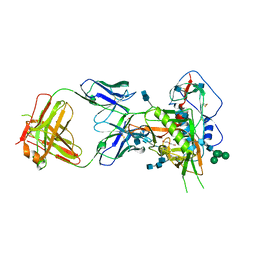 | | Crystal structure of glycosylated 426c HIV-1 gp120 core G459C in complex with glVRC01 A60C heavy chain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weidle, C, Pancera, M, Stamatatos, L, Gray, M. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
7BGU
 
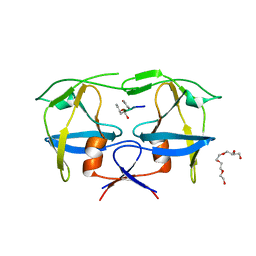 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
7BGT
 
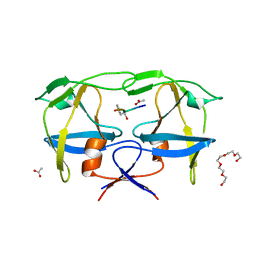 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | ACETATE ION, Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, ... | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
6S1U
 
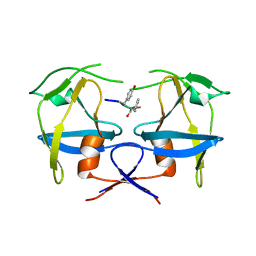 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
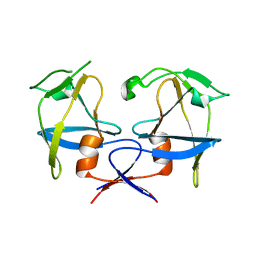 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
