3NC8
 
 | |
3NY4
 
 | | Crystal Structure of BlaC-K73A bound with Cefamandole | | Descriptor: | (6R,7R)-7-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-3-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
3N8R
 
 | |
3NDG
 
 | |
3N8L
 
 | |
3NDE
 
 | |
3NCK
 
 | |
3NBL
 
 | | Crystal Structure of BlaC-E166A covalently bound with Cefuroxime | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-[(1R)-1-{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}-2-oxoethyl]-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-06-03 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BlaC-E166A covalently bound with cephalosporins and penicillins
To be Published
|
|
3N7W
 
 | |
3N8S
 
 | | Crystal Structure of BlaC-E166A covalently bound with Cefamandole | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-2-oxoethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-11-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
1ENY
 
 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
3N6I
 
 | |
1ENZ
 
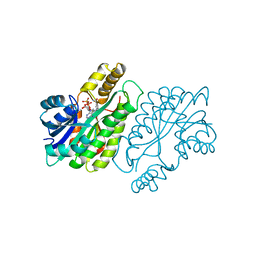 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
1BWZ
 
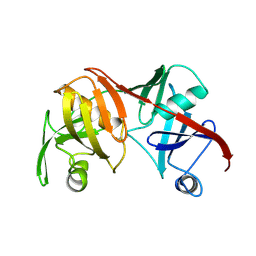 | |
1C3V
 
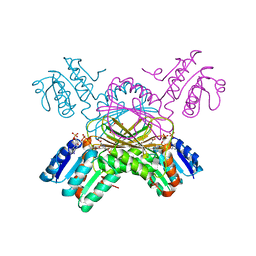 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
4HCX
 
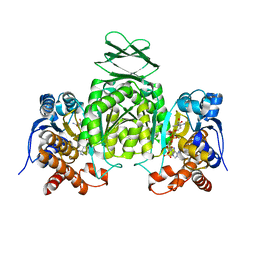 | |
6OVT
 
 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
4DF6
 
 | |
4X6T
 
 | | M.tuberculosis betalactamase complexed with inhibitor EC19 | | Descriptor: | 3-[(2R)-2-(dihydroxyboranyl)-2-{[(2R)-2-{[(4-ethyl-2,3-dioxo-3,4-dihydropyrazin-1(2H)-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibiting the beta-Lactamase of Mycobacterium tuberculosis (Mtb) with Novel Boronic Acid Transition-State Inhibitors (BATSIs).
ACS Infect Dis, 1, 2015
|
|
4Q8I
 
 | | Crystal Structure of beta-lactamase from M.tuberculosis covalently complexed with Tebipenem | | Descriptor: | (4R,5S)-3-(1-(4,5-dihydrothiazol-2-yl)azetidin-3-ylthio)-5-((2S,3R)-3-hydroxy-1-oxobutan-2-yl)-4-methyl-4,5- dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Blanchard, J. | | Deposit date: | 2014-04-27 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Tebipenem, a new carbapenem antibiotic, is a slow substrate that inhibits the beta-lactamase from Mycobacterium tuberculosis.
Biochemistry, 53, 2014
|
|
4QHC
 
 | | Structure of M.Tuberculosis Betalactamase (Blac) with inhibitor having novel mechanism | | Descriptor: | (3R,6R,7S)-7-[(2R,3aR)-hexahydropyrazolo[1,5-c][1,3]thiazin-2-yl]-6-(hydroxymethyl)-1,4-thiazepane-3-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Blanchard, J. | | Deposit date: | 2014-05-28 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Kinetic and Structural Characterization of the Interaction of 6-Methylidene Penem 2 with the beta-Lactamase from Mycobacterium tuberculosis.
Biochemistry, 54, 2015
|
|
4QB8
 
 | |
1BW9
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND PHENYLPYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYLPYRUVIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1BXG
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND BETA-PHENYLPROPIONATE | | Descriptor: | HYDROCINNAMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-05-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
4JLF
 
 | | Inhibitor resistant (R220A) substitution in the Mycobacterium tuberculosis beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Kurz, S, Blanchard, J, Bonomo, R. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis beta-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of beta-lactam-beta-lactamase inhibitor combinations.
Antimicrob.Agents Chemother., 57, 2013
|
|
