6BU1
 
 | |
7A78
 
 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6PI7
 
 | |
3OHW
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 721-860) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209E | | Descriptor: | Phycobilisome LCM core-membrane linker polypeptide | | Authors: | Kuzin, A, Su, M, Lew, S, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209E
To be Published
|
|
7A77
 
 | | Crystal structure of RXR alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7A79
 
 | | Crystal structure of RXR gamma LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | Nuclear receptor coactivator 2, PALMITIC ACID, Retinoic acid receptor RXR-gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
3MFX
 
 | | Crystal Structure of the sensory box domain of the sensory-box/GGDEF protein SO_1695 from Shewanella oneidensis, Northeast Structural Genomics Consortium Target SoR288B | | Descriptor: | Sensory box/GGDEF family protein | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-04 | | Release date: | 2010-04-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Northeast Structural Genomics Consortium Target SoR288B
To be Published
|
|
3P3Q
 
 | | Crystal Structure of MmoQ Response regulator from Methylococcus capsulatus str. Bath at the resolution 2.4A, Northeast Structural Genomics Consortium Target McR175M | | Descriptor: | MmoQ, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Abashidze, M, Seetharaman, J, Janjua, J, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-10-05 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR175M
To be Published
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3MTK
 
 | | X-Ray Structure of Diguanylate cyclase/phosphodiesterase from Caldicellulosiruptor saccharolyticus, Northeast Structural Genomics Consortium Target ClR27C | | Descriptor: | Diguanylate cyclase/phosphodiesterase | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Northeast Structural Genomics Consortium Target ClR27C
To be Published
|
|
4DRT
 
 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3PRU
 
 | | Crystal Structure of Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 1 (fragment 14-158) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR182A | | Descriptor: | CHLORIDE ION, Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, ... | | Authors: | Kuzin, A, Su, M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR182A
To be Published
|
|
4F2V
 
 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3NGW
 
 | | Crystal Structure of Molybdopterin-guanine dinucleotide biosynthesis protein A from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR189 | | Descriptor: | CALCIUM ION, Molybdopterin-guanine dinucleotide biosynthesis protein A (MobA) | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-13 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR189
To be Published
|
|
3OSJ
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 254-400) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | NITRATE ION, Phycobilisome LCM core-membrane linker polypeptide, SODIUM ION | | Authors: | Kuzin, A, Su, M, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-09 | | Release date: | 2010-10-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209C
To be published
|
|
4FGM
 
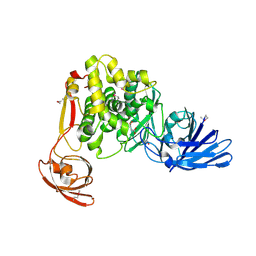 | | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis. Northeast Structural Genomics Consortium Target IlR60. | | Descriptor: | Aminopeptidase N family protein, MALEIC ACID, ZINC ION | | Authors: | Vorobiev, S, Su, M, Tong, T, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis.
To be Published
|
|
6NFT
 
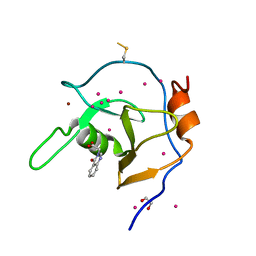 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (4-oxoquinazolin-3(4H)-yl)acetic acid | | Descriptor: | (4-oxoquinazolin-3(4H)-yl)acetic acid, 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Mann, M.K, Tempel, W, Bountra, C, Arrowmsmith, C.M, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
9BKS
 
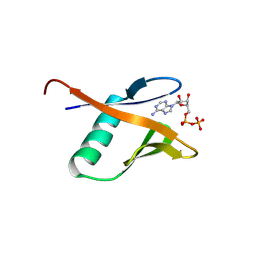 | | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of E3 ubiquitin-protein ligase TRIP12 | | Authors: | Kimani, S, Dong, A, Li, Y, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP
To be published
|
|
6ASB
 
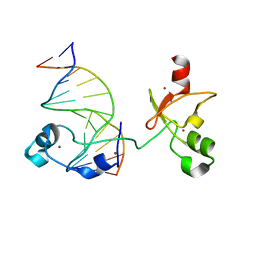 | | CXXC and PHD-type zinc finger regions of FBXL19 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), F-box/LRR-repeat protein 19, ZINC ION | | Authors: | Liu, K, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
3OBH
 
 | | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104 | | Descriptor: | ACETIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Abashidze, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-09-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | X-ray crystal structure of protein SP_0782 (7-79) from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
6B57
 
 | | tudor in complex with ligand | | Descriptor: | Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
6X9O
 
 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
8QLR
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR24 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-2-[1,3-bis(oxidanyl)propan-2-ylamino]-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
